













| General Information: |
|
| Name(s) found: |
FRS2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 807 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
response to red or far red light
[ISS]
|
| Molecular Function: |
zinc ion binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDDEDVEIDL LTKSSNVDVF CEASTSGNVA QCATVSELRN GMDFESKEAA YYFYREYARS 60
61 VGFGITIKAS RRSKRSGKFI DVKIACSRFG TKREKATAIN PRSCPKTGCK AGLHMKRKED 120
121 EKWVIYNFVK EHNHEICPDD FYVSVRGKNK PAGALAIKKG LQLALEEEDL KLLLEHFMEM 180
181 QDKQPGFFYA VDFDSDKRVR NVFWLDAKAK HDYCSFSDVV LFDTFYVRNG YRIPFAPFIG 240
241 VSHHRQYVLL GCALIGEVSE STYSWLFRTW LKAVGGQAPG VMITDQDKLL SDIVVEVFPD 300
301 VRHIFCLWSV LSKISEMLNP FVSQDDGFME SFGNCVASSW TDEHFERRWS NMIGKFELNE 360
361 NEWVQLLFRD RKKWVPHYFH GICLAGLSGP ERSGSIASHF DKYMNSEATF KDFFELYMKF 420
421 LQYRCDVEAK DDLEYQSKQP TLRSSLAFEK QLSLIYTDAA FKKFQAEVPG VVSCQLQKER 480
481 EDGTTAIFRI EDFEERQNFF VALNNELLDA CCSCHLFEYQ GFLCKHAILV LQSADVSRVP 540
541 SQYILKRWSK KGNNKEDKND KCATIDNRMA RFDDLCRRFV KLGVVASLSD EACKTALKLL 600
601 EETVKHCVSM DNSSKFPSEP DKLMTGGSIG LENEGVLDCA SKVSKKKKIQ KKRKVYCGPE 660
661 DATNRSEELR QETEQVSSRA PTFENCYIPQ ANMEEPELGS RATTLGVYYS TQQTNQGFPS 720
721 ISSIQNGYYG HPPTIQAMGN LHSIHERMSQ YETQPSMQGA FQGQTGFRGS AIRGCYDIEE 780
781 TLHDMTMGSS QFQGSDSSHP SDHRLSN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
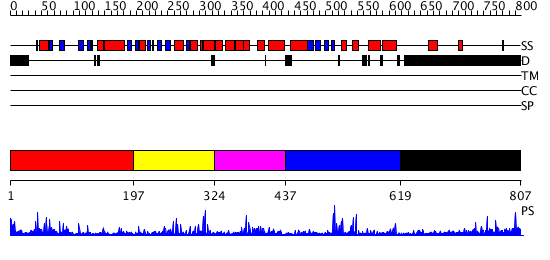
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..196] | 28.022276 | No description for PF03101.7 was found. No confident structure predictions are available. | |
| 2 | View Details | [197..323] | 29.455932 | No description for PF10551.1 was found. No confident structure predictions are available. | |
| 3 | View Details | [324..436] | 2.580992 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [437..618] | 4.408995 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [619..807] | 13.220997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)