













| General Information: |
|
| Name(s) found: |
FRS12_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 788 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
response to red or far red light
[ISS]
|
| Molecular Function: |
zinc ion binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MESVDTELTS FNNMVAKSSY PVRILHHNNG ISEDEEGGSG VEPYVGLEFD TAEEAREFYN 60
61 AYAARTGFKV RTGQLYRSRT DGTVSSRRFV CSKEGFQLNS RTGCTAFIRV QRRDTGKWVL 120
121 DQIQKEHNHE LGGEGSVEET TPRPSRAPAP TKLGVTVNPH RPKMKVVDES DRETRSCPGG 180
181 FKRFKGGGGE GEVSDDHHQT QQAKAVTGTE PYAGLEFGSA NEACQFYQAY AEVVGFRVRI 240
241 GQLFRSKVDG SITSRRFVCS REGFQHPSRM GCGAYMRIKR QDSGGWIVDR LNKDHNHDLE 300
301 PGKKNDAGMK KIPDDGTGGL DSVDLIELND FGNNHIKKTR ENRIGKEWYP LLLDYFQSRQ 360
361 TEDMGFFYAV ELDVNNGSCM SIFWADSRAR FACSQFGDSV VFDTSYRKGS YSVPFATIIG 420
421 FNHHRQPVLL GCAMVADESK EAFLWLFQTW LRAMSGRRPR SIVADQDLPI QQALVQVFPG 480
481 AHHRYSAWQI REKERENLIP FPSEFKYEYE KCIYQTQTIV EFDSVWSALI NKYGLRDDVW 540
541 LREIYEQREN WVPAYLRASF FAGIPINGTI EPFFGASLDA LTPLREFISR YEQALEQRRE 600
601 EERKEDFNSY NLQPFLQTKE PVEEQCRRLY TLTVFRIFQN ELVQSYNYLC LKTYEEGAIS 660
661 RFLVRKCGNE SEKHAVTFSA SNLNSSCSCQ MFEHEGLLCR HILKVFNLLD IRELPSRYIL 720
721 HRWTKNAEFG FVRDMESGVS AQDLKALMVW SLREAASKYI EFGTSSLEKY KLAYEIMREG 780
781 GKKLCWQR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
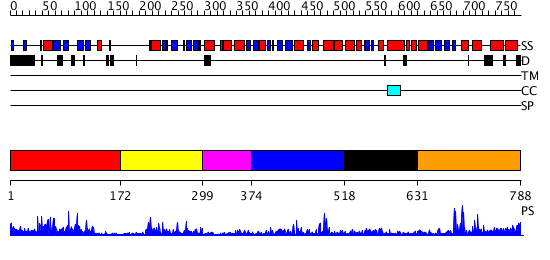
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..171] | 1.02 | No description for 2aydA was found. | |
| 2 | View Details | [172..298] | 3.465996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [299..373] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [374..517] | 18.704995 | View MSA. Confident ab initio structure predictions are available. | |
| 5 | View Details | [518..630] | 2.671996 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [631..788] | 6.373994 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)