













| General Information: |
|
| Name(s) found: |
gi|24376174
[NCBI NR]
gi|24351219 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 451 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
glutathione biosynthetic process
[ISS |
| Molecular Function: |
glutathione-disulfide reductase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAQHFDYICL GAGSGGIASA NRAAMRGAKV LLIEAKHVGG TCVNVGCVPK KVMWYGAHIA 60
61 EAMNLYAKDY GFDVSVNKFD WNTLVNSREA YIGRIHEAYG RGFTNNKVTL LNGYGRFVNG 120
121 NTIEVNGEHY TADHILIATG GAPTIPNIPG AEYGIDSDGF FALREQPKRV AVVGAGYIAV 180
181 EVAGVLHALG SETHLFVRKH APLRNFDPML IDALVDAMKT EGPTLHTNSV PQSVVKNADD 240
241 SLTLNLENGE SVTVDCLIWA IGRSPATGNI GLENTEVQLD SKGYVITDAQ QNTTHKGIYC 300
301 VGDIMAGGVE LTPVAVKAGR LLSERLFNAM SDAKMDYSQI PTVVFSHPPI GTMGLTEPEA 360
361 RAQYGDGNVK VYTSSFTSMY TAVTSHRQAC KMKLVCAGKE DKVVGIHGIG FGMDEILQGF 420
421 GVAMKMGATK ADFDAVVAIH PTGAEEFVTM R |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
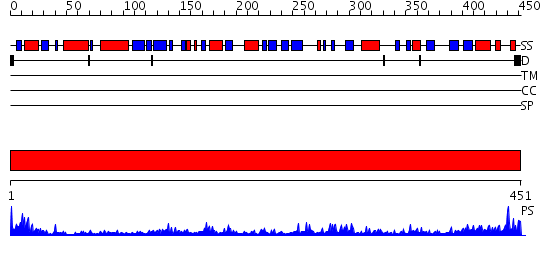
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..451] | 94.39794 | Glutathione reductase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.55 |
Source: Reynolds et al. (2008)