













| General Information: |
|
| Name(s) found: |
FRS1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 687 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
response to red or far red light
[ISS]
|
| Molecular Function: |
zinc ion binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSGECSNVQ LDDHRKNNLE IDEGREFESK EEAFEFYKEY ANSVGFTTII KASRRSRMTG 60
61 KFIDAKFVCT RYGSKKEDID TGLGTDGFNI PQARKRGRIN RSSSKTDCKA FLHVKRRQDG 120
121 RWVVRSLVKE HNHEIFTGQA DSLRELSGRR KLEKLNGAIV KEVKSRKLED GDVERLLNFF 180
181 TDMQVENPFF FYSIDLSEEQ SLRNIFWVDA KAMHGCRPRV ILTKHDQMLK EAVLEVFPSS 240
241 RHCFYMWDTL GQMPEKLGHV IRLEKKLVDE INDAIYGSCQ SEDFEKNWWE VVDRFHMRDN 300
301 VWLQSLYEDR EYWVPVYMKD VSLAGMCTAQ RSDSVNSGLD KYIQRKTTFK AFLEQYKKMI 360
361 QERYEEEEKS EIETLYKQPG LKSPSPFGKQ MAEVYTREMF KKFQVEVLGG VACHPKKESE 420
421 EDGVNKRTFR VQDYEQNRSF VVVWNSESSE VVCSCRLFEL KGFLCRHAMI VLQMSGELSI 480
481 PSQYVLKRWT KDAKSREVME SDQTDVESTK AQRYKDLCLR SLKLSEEASL SEESYNAVVN 540
541 VLNEALRKWE NKSNLIQNLE ESESVTAQDL PIHEEQNNTY DMNKDDNVAD TGQEYSLQEV 600
601 WKVTALQEQR NRYSILDDYL SAQHMSHEMG QINSMASNRN GYCSVHQNIH SLQGQSITHP 660
661 RLYETEQSSF RPEAMYERLQ DMVKDLN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
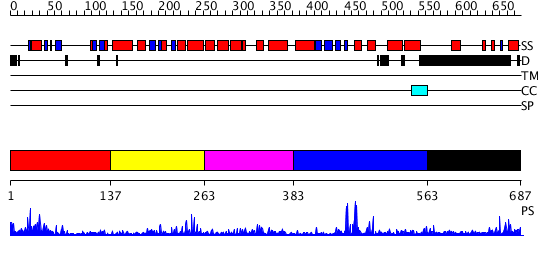
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..136] | 38.638272 | No description for PF03101.7 was found. No confident structure predictions are available. | |
| 2 | View Details | [137..262] | 4.040959 | No description for PF10551.1 was found. No confident structure predictions are available. | |
| 3 | View Details | [263..382] | 9.715996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [383..562] | 3.454994 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [563..687] | 3.345994 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)