













| General Information: |
|
| Name(s) found: |
DPYL2_PONPY
[Swiss-Prot]
|
| Description(s) found:
Found 3 descriptions. SHOW ALL |
|
| Organism: | Pongo pygmaeus |
| Length: | 572 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSYQGKKNIP RITSDRLLIK GGKIVNDDQS FYADIYMEDG LIKQIGENLI VPGGVKTIEA 60
61 HSRMVIPGGI DVHTRFQMPD QGMTSADDFF QGTKAALAGG TTMIIDHVVP EPGTSLLAAF 120
121 DQWREWADSK SCCDYSLHVD ISEWHKGIQE EMEALVKDHG VNSFLVYMAF KDRFQLTDCQ 180
181 IYEVLSVIRD IGAIAQVHAE NGDIIAEEQQ RILDLGITGP EGHVLSRPEE VEAEAVNRAI 240
241 TIANQTNCPL YITKVMSKSS AEVIAQARKK GTVVYGEPIT ASLGTDGSHY WSKNWAKAAA 300
301 FVTSPPLSPD PTTPDFLNSL LSCGDLQVTG SAHCTFNTAQ KAVGKDNFTL IPEGTNGTEE 360
361 RMSVIWDKAV VTGKMDENQF VAVTSTNAAK VFNLYPRKGR IAVGSDADLV IWDPDSVKTI 420
421 SAKTHNSSLE YNIFEGMECR GSPLVVISQG KIVLEDGTLH VTEGSGRYIP RKPFPDFVYK 480
481 RIKARSRLAE LRGVPRGLYD GPVCEVSVTP KTVTPASSAK TSPAKQQAPP VRNLHQSGFS 540
541 LSGAQIDDNI PRRTTQRIVA PPGGRANITS LG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
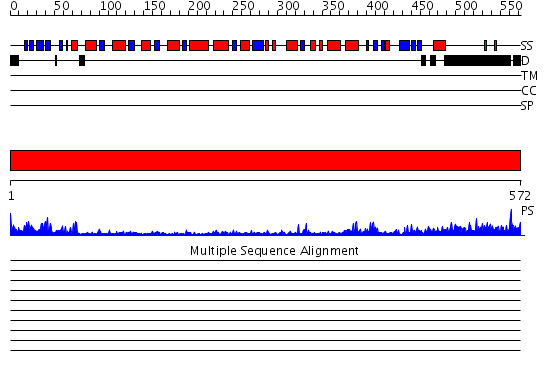
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..572] | 102.0 | X-ray structure of NYSGRC target T-45 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)