













| General Information: |
|
| Name(s) found: |
gi|537020
[NCBI NR]
gi|1361201 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 827 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTKIFIRDDN GGTEMSQDPF QEREAEKYAN PIPSREFILE HLTKREKPAS RDELAVELHI 60
61 EGEEQLEGLR RRLRAMERDG QLVFTRRQCY ALPERLDLVK GTVIGHRDGY GFLRVEGRKD 120
121 DLYLSSEQMK TCIHGDQVLA QPLGADRKGR REARIVRVLV PKTSQIVGRY FTEAGVGFVV 180
181 PDDSRLSFDI LIPPDQIMGA RMGFVVVVEL TQRPTRRTKA VGKIVEVLGD NMGTGMAVDI 240
241 ALRTHEIPYI WPQAVEQQVA GLKEEVPEEA KAGRVDLRDL PLVTIDGEDA RDFDDAVYCE 300
301 KKRGGGWRLW VAIADVSYYV RPSTPLDREA RNRGTSVYFP SQVIPMLPEV LSNGLCSLNP 360
361 QVDRLCMVCE MTVSSKGRLT GYKFYEAVMS SHARLTYTKV WHILQGDQDL REQYAPLVKH 420
421 LEELHNLYKV LDKAREERGG ISFESEEAKF IFNAERRIER IEQTQRNDAH KLIEECMILA 480
481 NISAARFVEK AKEPALFRIH DKPSTEAITS FRSVLAELGL ELPGGNKPEP RDYAELLESV 540
541 ADRPDAEMLQ TMLLRSMKQA IYDPENRGHF GLALQSYAHF TSPIRRYPDL TLHRAIKYLL 600
601 AKEQGHQGNT TETGGYHYSM EEMLQLGQHC SMAERRADEA TRDVADWLKC DFMLDQVGNV 660
661 FKGVISSVTG FGFFVRLDDL FIDGLVHVSS LDNDYYRFDQ VGQRLMGESS GQTYRLGDRV 720
721 EVRVEAVNMD ERKIDFSLIS SERAPRNVGK TAREKAKKGD AGKKGGKRRQ VGKKVNFEPD 780
781 SAFRGEKKTK PKAAKKDARK AKKPSAKTQK IAAATKAKRA AKKKVAE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
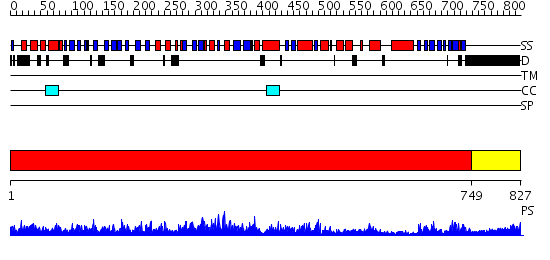
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..748] | 137.0 | No description for 2id0A was found. | |
| 2 | View Details | [749..827] | 24.522879 | No description for 2oceA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)