













| General Information: |
|
| Name(s) found: |
RSMA_ERWCT
[Swiss-Prot]
KSGA_ERWCT [Swiss-Prot] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Pectobacterium atrosepticum |
| Length: | 272 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNNRVHQGHF ARKRFGQNFL NDHFVIDSIV SAIHPQPGQA VVEIGPGLGA LTAPIGERMD 60
61 RFTVIELDRD LAARLEKHPT LKDKLTIIQQ DAMTIDFAAL AEQAGQPLRV FGNLPYNIST 120
121 PLMFHLFTYT QSIRDMHFML QKEVVNRLVA GPNSKTFGRL SVMAQYYCQI IPVLEVPPEA 180
181 FKPAPKVDSA VVRLVPHAEL PYPVSDIRML SRITTEAFNQ RRKTLRNSLG NLFTPETLTE 240
241 LGINITSRAE NVTVEQYCRL ANWLSEHPAK QE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
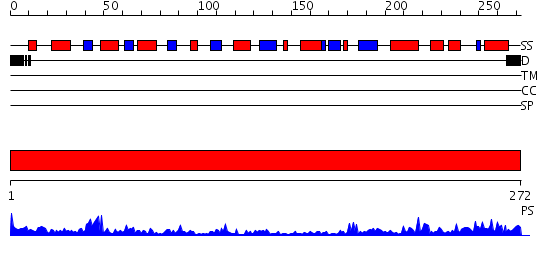
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..272] | 54.39794 | 2.1 Angstrom Crystal structure of KsgA: A Universally Conserved Adenosine Dimethyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)