













| General Information: |
|
| Name(s) found: |
gi|57237465
[NCBI NR]
gi|57166269 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Campylobacter jejuni RM1221 |
| Length: | 241 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane fumarate reductase complex
[ISS |
| Biological Process: |
anaerobic respiration
[ISS |
| Molecular Function: |
electron carrier activity
[ISS succinate dehydrogenase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRKLTIKAF KYNPLSKISK PHFVTYELEE TPFMTVFVCL TLIREKMDAD LSFDFVCRAG 60
61 ICGSCAMMIN GVPKLACKTL TKDYPDGVIE LMPMPAFRHI KDLSVNTGEW FEDMCKRVES 120
121 WVHNEKETDI SKLEERIEPE VADETFELDR CIECGICVAS CATKLMRPNF IAATGLLRTA 180
181 RYLQDPHDHR SVEDFYELVG DDDGVFGCMS LLACEDNCPK ELPLQSKIAY MRRQLVAQRN 240
241 K |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
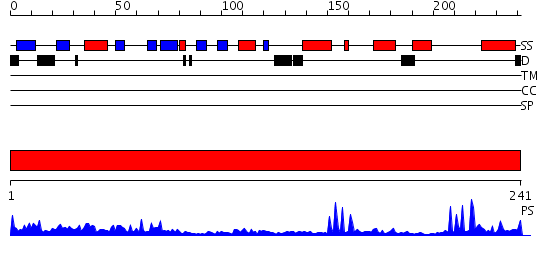
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..241] | 48.09691 | Fumarate reductase; Fumarate reductase iron-sulfur protein, N-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)