













| General Information: |
|
| Name(s) found: |
sv-PE /
FBpp0099748
[FlyBase]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 794 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[NAS]
|
| Biological Process: |
muscle organ development
[IMP]
neuron development [IMP] regulation of transcription [IEA][NAS] compound eye cone cell fate commitment [TAS] dendrite morphogenesis [IMP] |
| Molecular Function: |
DNA binding
[IEA]
transcription factor activity [TAS] RNA polymerase II transcription factor activity [NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLIMDIQTST HHIHGLGTHE LQHRILHPHI LHSTQEETLN TSTGQLEHDS QHLQQHHLTH 60
61 HQQQDVSPTL HNLQNTRTGD SLSTTINTNQ NQHGHQHLSG SNMYTSSQME DKSKANKYDE 120
121 YSSRTLSNIS DANTTPSANN FITQSQGIEW ITAMNDIQNG AEDSHSSQGS ISGDGHGGVN 180
181 QLGGVFVNGR PLPDVVRQRI VELAHNGVRP CDISRQLRVS HGCVSKILSR YYETGSFKAG 240
241 VIGGSKPKVA TPPVVDAIAN YKRENPTMFA WEIRDRLLAE AICSQDNVPS VSSINRIVRN 300
301 KAAEKAKHVH HHQQHHVSQS LGGGHIATES VDSSTGTIGE PQPPTSNSSA NSVNTNVSAS 360
361 ASVHASIPTS GTDSVQVSVG HINANSNETT HINSTAEQRT TGYSINGILG IQHGHHSHNN 420
421 NNNSSVNNNN NTESSCKRKR IEAHDENHDT NIHSDNDDGK RQRMSTYSGD QLYTNIWSGK 480
481 WCIKDDHKLL AELGNLTAST GNCPATYYEA SNGFSTTPIS GSGATASGND TSMLYDSITT 540
541 ISQTQSSIYT PAIGPSIGTG SLTPLVPISM HEMKLSANSI QEQTVPPFYT ALAFDGNYTS 600
601 MTSLENCSSL VGQEHIVMPE SSDSNTLCPI STRIVPDITE TNSTRVKEPL TNSDGCSEDN 660
661 NKEPEKSNSS QSSDHIASPH LHHIGEEQLR GNRRSNLNAS THPSSLIPLQ PSGGSSLLNI 720
721 NPSENRSDVE LNLSNNVGLY STPTVLPSFN HYSAGFLYFQ AAAQLYPVQI TPTILHIPSM 780
781 VELTDHMVTA LAVA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
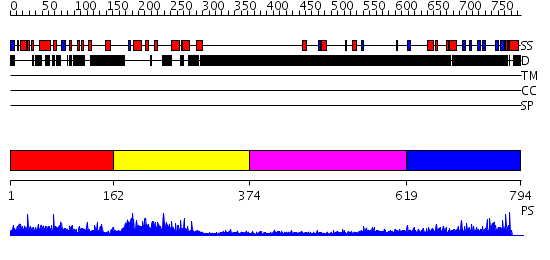
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..161] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [162..373] | 64.045757 | Pax-5 | |
| 3 | View Details | [374..618] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [619..794] | 5.243993 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)