













| General Information: |
|
| Name(s) found: |
YAT1 /
YAR035W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 687 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
alcohol metabolic process
[IGI carnitine metabolic process [IMP |
| Molecular Function: |
carnitine O-acetyltransferase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPNLKRLPIP PLQDTLNRYL ARVEPLQDER QNRRTRRTVL SAENLDALNT LHERLLEYDA 60
61 RLAESNPESS YIEQFWYDAY LLYDATVVLN VNPYFQLQDD PTIKDTPETA AQGPYGAHTV 120
121 QVRRAARLTT SILKFIRQIR HGTLRTDTVR GKTPLSMDQY ERLFGSSRIP PGPGEPSCHL 180
181 QTDATSHHVV AMYRGQFYWF DVLDTRNEPI FATPEQLEWN LYSIIMDAES AGSGSAPFGV 240
241 FTTESRRVWS NIRDYLFHAD DCTNWRNLKL IDSALFVVCL DDVAFAADQQ DELTRSMLCG 300
301 TSTINLDPHQ HQPPLNVQTG TCLNRWYDKL QLIVTKNGKA GINFEHTGVD GHTVLRLATD 360
361 IYTDSILSFA RGVTKNVVDI FSDDDGKPSS SSLASAAHSA NLITIPRKLE WRTDNFLQSS 420
421 LHFAETRISD LISQYEFVNL DFSNYGASHI KTVFKCSPDA FVQQVFQVAY FALYGRFETV 480
481 YEPAMTKAFQ NGRTEAIRSV TGQSKLFVKS LLDQDASDAT KIQLLHDACT AHSQITRECS 540
541 QGLGQDRHLY ALYCLWNQWY KDKLELPPIF RDKSWTTMQN NVLSTSNCGN PCLKSFGFGP 600
601 VTANGFGIGY IIRDHSVSVV VSSRHRQTAR FASLMEKSLL EIDRIFKRQQ ARAAKPAART 660
661 TASANTKSED MKYLLSGYDY FDVSVSG |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |

|
CAT2 YAT1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Hazbun TR, et al. (2003) |
New Feature: Upload Your Own Microscopy Data
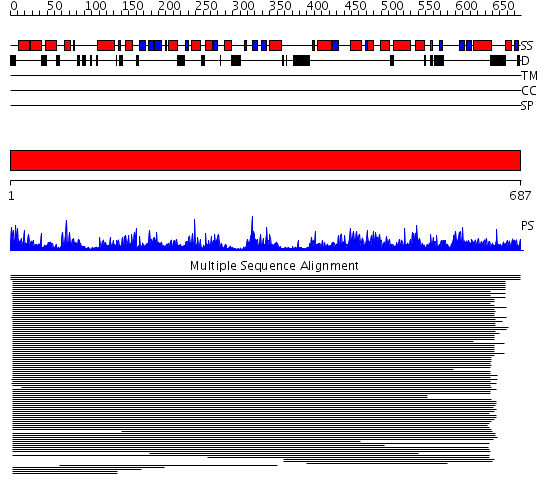
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..687] | 294.886057 | Choline/Carnitine o-acyltransferase No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)