













| General Information: |
|
| Name(s) found: |
PBX3
[HGNC (HUGO)]
|
| Description(s) found:
Found 33 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 434 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDDQSRMLQT LAGVNLAGHS VQGGMALPPP PHGHEGADGD GRKQDIGDIL HQIMTITDQS 60
61 LDEAQAKKHA LNCHRMKPAL FSVLCEIKEK TGLSIRGAQE EDPPDPQLMR LDNMLLAEGV 120
121 SGPEKGGGSA AAAAAAAASG GSSDNSIEHS DYRAKLTQIR QIYHTELEKY EQACNEFTTH 180
181 VMNLLREQSR TRPISPKEIE RMVGIIHRKF SSIQMQLKQS TCEAVMILRS RFLDARRKRR 240
241 NFSKQATEIL NEYFYSHLSN PYPSEEAKEE LAKKCSITVS QVSNWFGNKR IRYKKNIGKF 300
301 QEEANLYAAK TAVTAAHAVA AAVQNNQTNS PTTPNSGSSG SFNLPNSGDM FMNMQSLNGD 360
361 SYQGSQVGAN VQSQVDTLRH VINQTGGYSD GLGGNSLYSP HNLNANGGWQ DATTPSSVTS 420
421 PTEGPGSVHS DTSN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
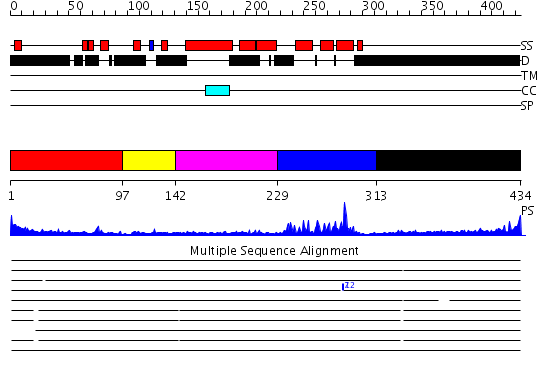
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..96] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [97..141] | 1.07 | No description for 2h8rA was found. | |
| 3 | View Details | [142..228] | 9.0 | THE SOLUTION STRUCTURE OF THE OCT-1 POU-SPECIFIC DOMAIN REVEALS A STRIKING SIMILARITY TO THE BACTERIOPHAGE LAMBDA REPRESSOR DNA-BINDING DOMAIN | |
| 4 | View Details | [229..312] | 18.09691 | pbx1 | |
| 5 | View Details | [313..434] | 1.768992 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)