













| General Information: |
|
| Name(s) found: |
gi|123999428
[NCBI NR]
gi|123991812 [NCBI NR] |
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | synthetic construct |
| Length: | 835 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAELSEEALL SVLPTIRVPK AGDRVHKDEC AFSFDTPESE GGLYICMNTF LGFGKQYVER 60
61 HFNKTGQRVY LHLRRTRRPK EEDPATGTGD PPRKKPTRLA IGVEGGFDLS EEKFELDEDV 120
121 KIVILPDYLE IARDGLGGLP DIVRDRVTSA VEALLSADSA SRKQEVQAWD GEVRQVSKHA 180
181 FSLKQLDNPA RIPPCGWKCS KCDMRENLWL NLTDGSILCG RRYFDGSGGN NHAVEHYRET 240
241 GYPLAVKLGT ITPDGADVYS YDEDDMVLDP SLAEHLSHFG IDMLKMQKTD KTMTELEIDM 300
301 NQRIGEWELI QESGVPLKPL FGPGYTGIRN LGNSCYLNSV VQVLFSIPDF QRKYVDKLEK 360
361 IFQNAPTDPT QDFSTQVAKL GHGLLSGEYS KPVPESGDGE RVPEQKEVQD GIAPRMFKAL 420
421 IGKGHPEFST NRQQDAQEFF LHLINMVERN CRSSENPNEV FRFLVEEKIK CLATEKVKYT 480
481 QRVDYIMQLP VPMDAALNKE ELLEYEEKKR QAEEEKMALP ELVRAQVPFS SCLEAYGAPE 540
541 QVDDFWSTAL QAKSVAVKTT RFASFPDYLV IQIKKFTFGL DWVPKKLDVS IEMPEELDIS 600
601 QLRGTGLQPG EEELPDIAPP LVTPDEPKAP MLDESVIIQL VEMGFPMDAC RKAVYYTGNS 660
661 GAEAAMNWVM SHMDDPDFAN PLILPGSSGP GSTSAAADPP PEDCVTTIVS MGFSRDQALK 720
721 ALRATNNSLE RAVDWIFSHI DDLDAEAAMD ISEGRSAADS ISESVPVGPK VRDGPGKYQL 780
781 FAFISHMGTS TMCGHYVCHI KKEGRWVIYN DQKVCASEKP PKDLGYIYFY QRVAS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
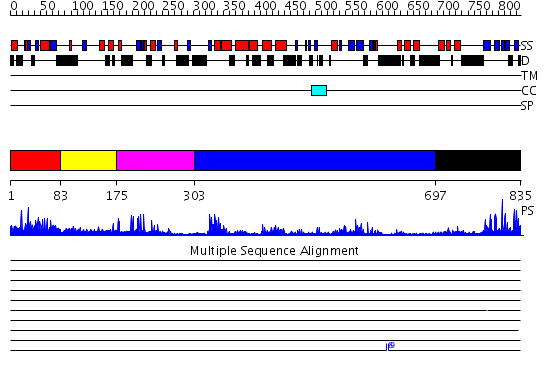
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] | 2.01 | No description for 2idaA was found. | |
| 2 | View Details | [83..174] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [175..302] | 26.522879 | No description for 2g43A was found. | |
| 4 | View Details | [303..696] | 51.0 | No description for 2ibiA was found. | |
| 5 | View Details | [697..835] | 4.11 | Crystal structure of HAUSP |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)