













| General Information: |
|
| Name(s) found: |
CG7759-PA /
FBpp0087232
[FlyBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 660 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytoplasm [IDA] |
| Biological Process: | NONE FOUND |
| Molecular Function: |
zinc ion binding
[IEA]
transcription repressor activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKTPLSQKTG FFAEYYLKLK DQAGPNCESD IAKLSACKSD EERVAYVEKL PWVQAGDANL 60
61 VVNQEFAGKN ASLAAEIKER ATSAFKAKKW LEAMMLYTRS YVALPSENVA EIRVVLANRS 120
121 ATLYHMQKYQ ECLIDIKRAL DLSYSKDLIY KLYERQARCY MALKDYPHTI DSFKKCITAM 180
181 DDSTLASDKR AKLNLDAMTM IKMLQNDPRT AKQEAKQQKQ KIALDQAKPV KLENEFVSPL 240
241 VRIDSNRQEG RFARASADVK PGEELLVERP FVSVLLEKFA KTHCENCFMR TVVPVACPRC 300
301 ADVLYCSEQC REEASKKYHK YECGIVPIIW RSGASINNHI ALRIIASKPL DYFLKLKPTI 360
361 DEELTPEQLI SLPKDDFRRV AQLERHQGER QPSNFFQHVL MARFLTNCLR AGGYFGSEPK 420
421 PDEVSIICSL VLRSLQFIQF NTHEVAELHK FSSSGREKSI FIGGAIYPTL ALFNHSCDPG 480
481 VVRYFRGTTI HINSVRPIEA GLPINENYGP MYTQDERSER QARLKDLYWF ECSCDACIDN 540
541 WPKFDDLPRD VIRFRCDAPN NCSAVIEVPP SCNDFMVKCV TCGEITNILK GLKVMQDTEM 600
601 MTRTAKRLYE TGEYPKALAK FVDLIRIMYE VLAPPFPDFC ESQQHLKDCF LNLGNVYTLD 660
661 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
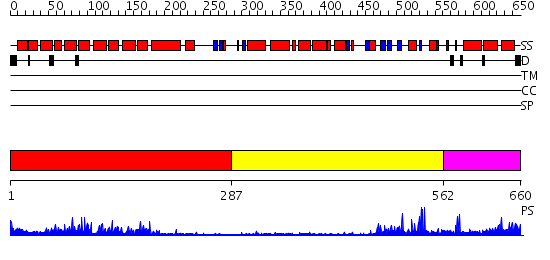
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..286] | 28.30103 | The superhelical TPR domain of O-linked GlcNAc transferase reveals structural similarities to importin alpha. | |
| 2 | View Details | [287..561] | 5.69897 | No description for 2o8jA was found. | |
| 3 | View Details | [562..660] | 0.987 | Crystal Structure of FKBP52 C-terminal Domain complex with the C-terminal peptide MEEVD of Hsp90 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)