













| General Information: |
|
| Name(s) found: |
aur-PA /
FBpp0081957
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 411 amino acids |
Gene Ontology: |
|
| Cellular Component: |
centrosome
[NAS]
|
| Biological Process: |
centrosome separation
[IMP]
centrosome cycle [IMP] asymmetric protein localization involved in cell fate determination [IMP] establishment of spindle orientation [IMP] mitotic centrosome separation [IMP] protein amino acid phosphorylation [IEA][NAS] regulation of neurogenesis [IMP] microtubule-based process [TAS] asymmetric protein localization [IMP] |
| Molecular Function: |
ATP binding
[IEA]
protein kinase activity [NAS] protein serine/threonine kinase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSHPSDHVLR PKENAPHRMP EKSAAVHNMQ KNLLLGKKPN SENMAPASKP LPGSSGALIR 60
61 KPGLGGSNSI ASSEGNNFQK PMVPSVKKTT SEFAAPAPVA PIKKPESLSK QKPTAASSES 120
121 SKELGAASSS AEKEKTKTET QPQKPKKTWE LNNFDIGRLL GRGKFGNVYL AREKESQFVV 180
181 ALKVLFKRQI GESNVEHQVR REIEIQSHLR HPHILRLYAY FHDDVRIYLI LEYAPQGTLF 240
241 NALQAQPMKR FDERQSATYI QALCSALLYL HERDIIHRDI KPENLLLGHK GVLKIADFGW 300
301 SVHEPNSMRM TLCGTVDYLP PEMVQGKPHT KNVDLWSLGV LCFELLVGHA PFYSKNYDET 360
361 YKKILKVDYK LPEHISKAAS HLISKLLVLN PQHRLPLDQV MVHPWILAHT Q |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
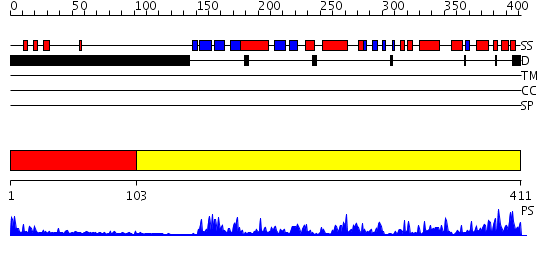
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..102] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [103..411] | 74.30103 | No description for 2j4zA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)