













| General Information: |
|
| Name(s) found: |
ssr1 /
SPAC17G6.10
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 527 amino acids |
Gene Ontology: |
|
| Cellular Component: |
SWI/SNF complex
[IDA nucleus [IDA nuclear chromatin [IC cytosol [IDA RSC complex [IDA |
| Biological Process: |
chromatin remodeling
[ISS positive regulation of gene-specific transcription from RNA polymerase II promoter [NAS negative regulation of gene-specific transcription from RNA polymerase II promoter [NAS |
| Molecular Function: |
DNA binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNVTENKHD LHENGVPDSA QPMELDVSKK EDTEPEVRDE AKEFLLSQLP QVEVPEWAQW 60
61 FDFSKVHEIE KKQNPEFFDG KNTSKTPEVY KEYRDFMIST FRLNSKVYLT FTACRRNLAG 120
121 DVCAVLRVHR FLEQWGLINY NVNPDTRPSK IGPPSTSHFQ ILADTPRGLV PLLPPPSSSI 180
181 PRSKAVTIED PSIVRTNIYD PSLDDVLKGK GSTPNQKPSL SNLHENNIDQ SDSPQHCYCC 240
241 GNKFNESYYQ SQTAQKYNVC ISCYQQNRFP SPTTIADYKE VAIQNKIEDD DTWTAQELVL 300
301 LSEGVEMYSD DWAKVASHVN TKSVEECILK FLNLPSSDKA LFKMDKVHTN PVVDSLQGKN 360
361 PILSVVSFLA KMVPPSSFTQ KSSAKEEESD KVKGESVYPK PESESYDVEM NGKSLEDSDS 420
421 LSELYLTNEE KKMASIIKDS VNVQIKLIES KLSHFDYLDQ HIRLKSQELD AFAQATYREK 480
481 LYMKRECQNA RKKIEQQLQS KDTAANGLPV QSSAETSVIP ASSMSPS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
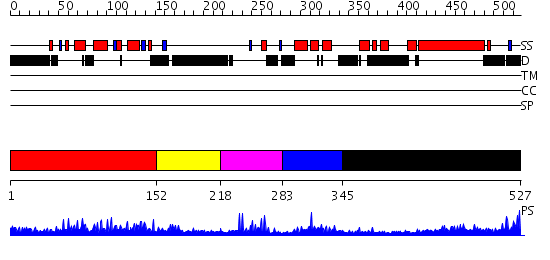
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..151] | 34.0 | Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes | |
| 2 | View Details | [152..217] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [218..282] | 12.69897 | No description for 2e5rA was found. | |
| 4 | View Details | [283..344] | 2.94 | Solution structure of the SANT domain of human KIAA1915 protein | |
| 5 | View Details | [345..527] | 3.522879 | Crystal Structure Analysis of ClpB |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)