













| General Information: |
|
| Name(s) found: |
mlt-9 /
CE23633
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 573 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA |
| Biological Process: |
molting cycle, protein-based cuticle
[IMP locomotory behavior [IMP locomotion [IMP nematode larval development [IMP positive regulation of growth rate [IMP growth [IMP |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRSATILLLL VHGTLACFPT SQRSYQYGAP TQQFAACTPQ CCGCSQAQPG PQPAAAAAAR 60
61 VAAAGKTPTE PISVQGSSIE GLLGQKVSLT EFTADGYNAE GGGPVRSGNA LTCNFDGRPC 120
121 CWANVPPPDD QLDWQIASGV PVQLQNRGVP QPEGSYLIAY AKSAAPSDEA QFASCSIGCA 180
181 SSDIVVRAKH WQSESVLLQV CLRESFPSNA ERNPLINCQE FPYVDGMGTT EVILPKTSLV 240
241 DIVFVASNFI GDQGDVAILD DIEVSYDRNG SECQNEFEEG AEEEDNQDNE NAIRAEEIEA 300
301 AAKNKRRELF EKTQPEGKIV SATERFGANG FRGGEKAFRG GAETGAGAGA GSKFGGSFEG 360
361 AENGNAAVGT SHTEETVFAS VKTAKLPSGH VVAKGAKTAE FSEVVCNATK CDFENENTCG 420
421 YKDAHTTQSI RGLTTKFNVV TGQFMNRVTG IKESTEGEYY AATFLFPREM AGLEASVAPF 480
481 TEQSRIRFHY YEGTHGVQLK GCCTNIENCP FSSDKFVTVA DRAWKTASFK CPKGTDKVIF 540
541 ICENTRTNQG ACAVDGIGLV ENEGPLKTAK PLC |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
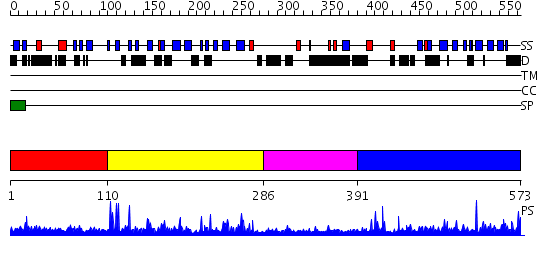
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..109] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [110..285] | 15.522879 | Crystal structure of the MAM-Ig module of receptor protein tyrosine phosphatase mu | |
| 3 | View Details | [286..390] | 4.064997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [391..573] | 2.88 | No description for 2v5yA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|