













| General Information: |
|
| Name(s) found: |
CE31335
[WormBase]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 317 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
molting cycle, protein-based cuticle
[IMP hermaphrodite genitalia development [IMP locomotory behavior [IMP nematode larval development [IMP positive regulation of growth rate [IMP gamete generation [IMP protein amino acid phosphorylation [IEA growth [IMP |
| Molecular Function: |
ATP binding
[IEA protein tyrosine kinase activity [IEA zinc ion binding [IEA RNA binding [IEA protein kinase activity [IEA protein serine/threonine kinase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDPYQQGGRG GGFPARGGRG GGHGGGYPQE GYGAAAGGYG GYDPYNPYGA AGGYGMYPGQ 60
61 GYPPQEMTSP LDAEIQAVLR EIHLEVTGLE TSGDQFRNAR RLLNSEKERL ENNIDPEWLE 120
121 VDVAKPVKVC KKILVPIYRH PNFNFIGKVL GPKGATLQTL CKTHKCHIYI LGRGSTKDRE 180
181 KEAELLASGD PQHAHFSGPL HVKVETVAPA YIAYGRVAAV IEELSRILQP IHEDTTPAHL 240
241 KNGSGDGEEK EDEEKKEGGE GGGRGGRGGR GGFRGGFRGG RGGFGGPGGP MGGRGGMGGM 300
301 GGRGRGGQAA GVHNPIT |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
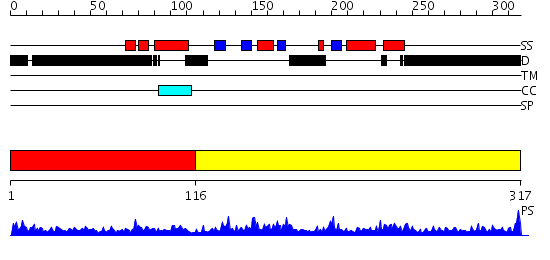
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..115] | 3.136993 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [116..317] | 46.69897 | Solution structure of the KH-QUA2 region of the Xenopus STAR-GSG Quaking protein. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 | No functions predicted. | ||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)