













| General Information: |
|
| Name(s) found: |
YMCB_BACSU
[Swiss-Prot]
MIAB_BACSU [Swiss-Prot] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Bacillus subtilis |
| Length: | 509 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNEKQKLESG QVHPSDKKSE KDYSKYFEAV YIPPSLKDAK KRGKEAVTYH NDFKISEQFK 60
61 GLGDGRKFYI RTYGCQMNEH DTEVMAGIFM ALGYEATNSV DDANVILLNT CAIRENAENK 120
121 VFGELGHLKA LKKNNPDLIL GVCGCMSQEE SVVNRILKKH PFVDMIFGTH NIHRLPELLS 180
181 EAYLSKEMVV EVWSKEGDVI ENLPKVRNGK IKGWVNIMYG CDKFCTYCIV PYTRGKERSR 240
241 RPEDIIQEVR RLASEGYKEI TLLGQNVNAY GKDFEDMTYG LGDLMDELRK IDIPRIRFTT 300
301 SHPRDFDDRL IEVLAKGGNL LDHIHLPVQS GSSEVLKLMA RKYDRERYME LVRKIKEAMP 360
361 NASLTTDIIV GFPNETDEQF EETLSLYREV EFDSAYTFIY SPREGTPAAK MKDNVPMRVK 420
421 KERLQRLNAL VNEISAKKMK EYEGKVVEVL VEGESKNNPD ILAGYTEKSK LVNFKGPKEA 480
481 IGKIVRVKIQ QAKTWSLDGE MVGEAIEVK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
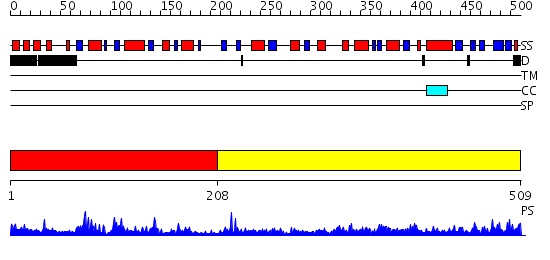
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..207] | 0.97 | GLUTAMATE MUTASE (B12-BINDING SUBUNIT), NMR, MINIMIZED AVERAGE STRUCTURE | |
| 2 | View Details | [208..509] | 61.69897 | No description for 2qgqA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 |
|
||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)