













| General Information: |
|
| Name(s) found: |
PSMD4_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 386 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[TAS]
proteasome regulatory particle, base subcomplex [IDA][ISS] membrane [IDA] cytosol [IDA] proteasome complex [IDA][TAS] |
| Biological Process: |
response to salt stress
[IMP]
protein catabolic process [TAS] leaf senescence [IMP] pollen development [IMP] root hair elongation [IMP] response to DNA damage stimulus [IMP] proteasome assembly [IMP] response to cytokinin stimulus [IEP][IMP] post-embryonic root development [IMP] [NOT]proteasome core complex assembly [IMP] ubiquitin-dependent protein catabolic process [TAS] response to auxin stimulus [IMP] response to sucrose stimulus [IMP] response to heat [IMP] proteasomal ubiquitin-dependent protein catabolic process [IMP] response to abscisic acid stimulus [IMP] stamen formation [IMP] leaf development [IMP] regulation of seed germination [IMP] response to misfolded protein [IMP] |
| Molecular Function: |
peptide receptor activity
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVLEATMICI DNSEWMRNGD YSPSRLQAQT EAVNLLCGAK TQSNPENTVG ILTMAGKGVR 60
61 VLTTPTSDLG KILACMHGLD VGGEINLTAA IQIAQLALKH RQNKNQRQRI IVFAGSPIKY 120
121 EKKALEIVGK RLKKNSVSLD IVNFGEDDDE EKPQKLEALL TAVNNNDGSH IVHVPSGANA 180
181 LSDVLLSTPV FTGDEGASGY VSAAAAAAAA GGDFDFGVDP NIDPELALAL RVSMEEERAR 240
241 QEAAAKKAAD EAGQKDKDGD TASASQETVA RTTDKNAEPM DEDSALLDQA IAMSVGDVNM 300
301 SEAADEDQDL ALALQMSMSG EESSEATGAG NNLLGNQAFI SSVLSSLPGV DPNDPAVKEL 360
361 LASLPDESKR TEEEESSSKK GEDEKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
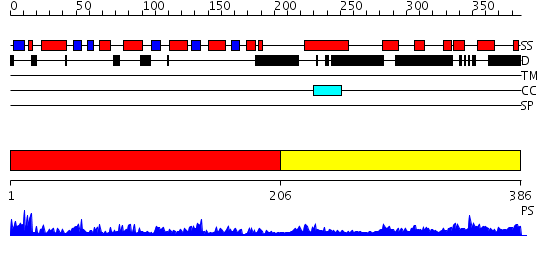
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..205] | 9.69897 | No description for 2i6qA was found. | |
| 2 | View Details | [206..386] | 14.69897 | Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)