













| General Information: |
|
| Name(s) found: |
FRS6_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 703 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
response to red or far red light
[ISS]
|
| Molecular Function: |
zinc ion binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MERSESVDED VQASAYLEND EVRERDDPMS DTLRIGEEAL VCSSEHEIDV GFCQNEERKS 60
61 EEETMGGFDE QSGLLVDERK EFDAPAVGME FESYDDAYNY YNCYASEVGF RVRVKNSWFK 120
121 RRSKEKYGAV LCCSSQGFKR INDVNRVRKE TRTGCPAMIR MRQVDSKRWR VVEVTLDHNH 180
181 LLGCKLYKSV KRKRKCVSSP VSDAKTIKLY RACVVDNGSN VNPNSTLNKK FQNSTGSPDL 240
241 LNLKRGDSAA IYNYFCRMQL TNPNFFYLMD VNDEGQLRNV FWADAFSKVS CSYFGDVIFI 300
301 DSSYISGKFE IPLVTFTGVN HHGKTTLLSC GFLAGETMES YHWLLKVWLS VMKRSPQTIV 360
361 TDRCKPLEAA ISQVFPRSHQ RFSLTHIMRK IPEKLGGLHN YDAVRKAFTK AVYETLKVVE 420
421 FEAAWGFMVH NFGVIENEWL RSLYEERAKW APVYLKDTFF AGIAAAHPGE TLKPFFERYV 480
481 HKQTPLKEFL DKYELALQKK HREETLSDIE SQTLNTAELK TKCSFETQLS RIYTRDMFKK 540
541 FQIEVEEMYS CFSTTQVHVD GPFVIFLVKE RVRGESSRRE IRDFEVLYNR SVGEVRCICS 600
601 CFNFYGYLCR HALCVLNFNG VEEIPLRYIL PRWRKDYKRL HFADNGLTGF VDGTDRVQWF 660
661 DQLYKNSLQV VEEGAVSLDH YKVAMQVLQE SLDKVHSVEE KQD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
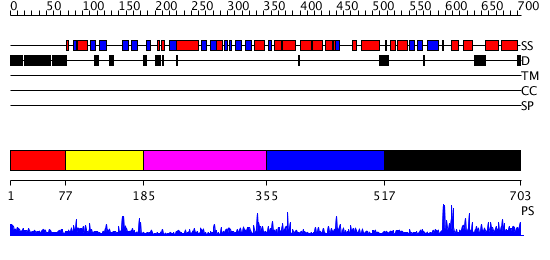
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..76] | 2.245996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [77..184] | 27.408935 | No description for PF03101.6 was found. No confident structure predictions are available. | |
| 3 | View Details | [185..354] | 1.671984 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [355..516] | 14.680992 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [517..703] | 5.305994 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)