













| General Information: |
|
| Name(s) found: |
FRS7_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 764 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
response to red or far red light
[ISS]
|
| Molecular Function: |
zinc ion binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVVKTYPLGM VGTNNGIAEN EGDSGLEPYV GLEFDTAEEA RDYYNSYATR TGFKVRTGQL 60
61 YRSRTDGTVS SRRFVCSKEG FQLNSRTGCP AFIRVQRRDT GKWVLDQIQK EHNHDLGGHI 120
121 EEAQTTPRPS VQQRAPAPTK LGISVPHRPK MKVVDEADKG RSCPSGVISF KRFKGAEDSD 180
181 GQTQPKATEP YAGLEFNSAN EACQFYQAYA EVVGFRVRIG QLFRSKVDGS ITSRRFVCSK 240
241 EGFQHPSRMG CGAYMRIKRQ DSGGWIVDRL NKDHNHDLEP GKKNAGMKKI TDDVTGGLDS 300
301 VDLIELNDLS NHISSTRENT IGKEWYPVLL DYFQSKQAED MGFFYAIELD SNGSCMSIFW 360
361 ADSRSRFACS QFGDAVVFDT SYRKGDYSVP FATFIGFNHH RQPVLLGGAL VADESKEAFS 420
421 WLFQTWLRAM SGRRPRSMVA DQDLPIQQAV AQVFPGTHHR FSAWQIRSKE RENLRSFPNE 480
481 FKYEYEKCLY QSQTTVEFDT MWSSLVNKYG LRDNMWLREI YEKREKWVPA YLRASFFGGI 540
541 HVDGTFDPFY GTSLNSLTSL REFISRYEQG LEQRREEERK EDFNSYNLQP FLQTKEPVEE 600
601 QCRRLYTLTI FRIFQSELAQ SYNYLGLKTY EEGAISRFLV RKCGNENEKH AVTFSASNLN 660
661 ASCSCQMFEY EGLLCRHILK VFNLLDIREL PSRYILHRWT KNAEFGFVRD VESGVTSQDL 720
721 KALMIWSLRE AASKYIEFGT SSLEKYKLAY EIMREGGKKL CWQR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
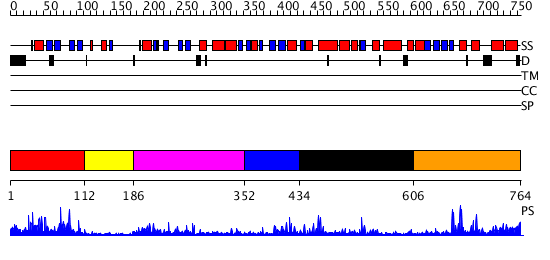
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..111] | 1.03 | No description for 2aydA was found. | |
| 2 | View Details | [112..185] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [186..351] | 36.09691 | No description for PF03101.6 was found. No confident structure predictions are available. | |
| 4 | View Details | [352..433] | N/A | Confident ab initio structure predictions are available. | |
| 5 | View Details | [434..605] | 18.717995 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [606..764] | 6.366995 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)