













| General Information: |
|
| Name(s) found: |
FRS3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 851 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
response to red or far red light
[ISS]
|
| Molecular Function: |
zinc ion binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDVHLVEENV SMGNHEIGDE GDVEPSDCSG QNNMDNSLGV QDEIGIAEPC VGMEFNSEKE 60
61 AKSFYDEYSR QLGFTSKLLP RTDGSVSVRE FVCSSSSKRS KRRLSESCDA MVRIELQGHE 120
121 KWVVTKFVKE HTHGLASSNM LHCLRPRRHF ANSEKSSYQE GVNVPSGMMY VSMDANSRGA 180
181 RNASMATNTK RTIGRDAHNL LEYFKRMQAE NPGFFYAVQL DEDNQMSNVF WADSRSRVAY 240
241 THFGDTVTLD TRYRCNQFRV PFAPFTGVNH HGQAILFGCA LILDESDTSF IWLFKTFLTA 300
301 MRDQPPVSLV TDQDRAIQIA AGQVFPGARH CINKWDVLRE GQEKLAHVCL AYPSFQVELY 360
361 NCINFTETIE EFESSWSSVI DKYDLGRHEW LNSLYNARAQ WVPVYFRDSF FAAVFPSQGY 420
421 SGSFFDGYVN QQTTLPMFFR LYERAMESWF EMEIEADLDT VNTPPVLKTP SPMENQAANL 480
481 FTRKIFGKFQ EELVETFAHT ANRIEDDGTT STFRVANFEN DNKAYIVTFC YPEMRANCSC 540
541 QMFEHSGILC RHVLTVFTVT NILTLPPHYI LRRWTRNAKS MVELDEHVSE NGHDSSIHRY 600
601 NHLCREAIKY AEEGAITAEA YNIALGQLRE GGKKVSVVRK RIGRAAPPSS HGGGIGSGDK 660
661 TSLSAADTTP LLWPRQDEMI RRFNLNDGGA RAQSVSDLNL PRMAPVSLHR DDTAPENMVA 720
721 LPCLKSLTWG MESKNTMPGG RVAVINLKLH DYRKFPSADM DVKFQLSSVT LEPMLRSMAY 780
781 ISEQLSSPAN RVAVINLKLQ DTETTTGESE VKFQVSRDTL GAMLRSMAYI REQLSIVGEL 840
841 QTESQAKKQR K |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
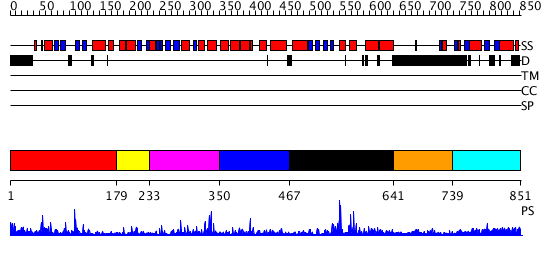
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..178] | 23.744727 | No description for PF03101.7 was found. No confident structure predictions are available. | |
| 2 | View Details | [179..232] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [233..349] | 26.60206 | No description for PF10551.1 was found. No confident structure predictions are available. | |
| 4 | View Details | [350..466] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [467..640] | 6.576994 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [641..738] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [739..851] | 2.137995 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)