













| General Information: |
|
| Name(s) found: |
EBSD_ENTFA
[Swiss-Prot]
AROD_ENTFA [Swiss-Prot] |
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Enterococcus faecalis |
| Length: | 253 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKPVIVKNVR IGEGNPKIVV PIVAPTAEDI LAEATASQTL DCDLVEWRLD YYENVADFSD 60
61 VCNLSQQVME RLGQKPLLLT FRTQKEGGEM AFSEENYFAL YHELVKKGAL DLLDIELFAN 120
121 PLAADTLIHE AKKAGIKIVL CNHDFQKTPS QEEIVARLRQ MQMRQADICK IAVMPQDATD 180
181 VLTLLSATNE MYTHYASVPI VTMSMGQLGM ISRVTGQLFG SALTFGSAQQ ASAPGQLSVQ 240
241 VLRNYLKTFE QNK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
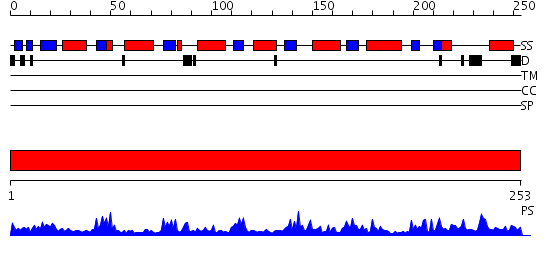
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..253] | 83.0 | Native 3-dehydroquinase from Salmonella typhi |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.79 |
Source: Reynolds et al. (2008)