













| Protein: | gi|223365863 |
| From Publication: | Noble WS, et al. (2009) Manuscript in preparation. |
| Complexes containing gi|223365863: | 7 |
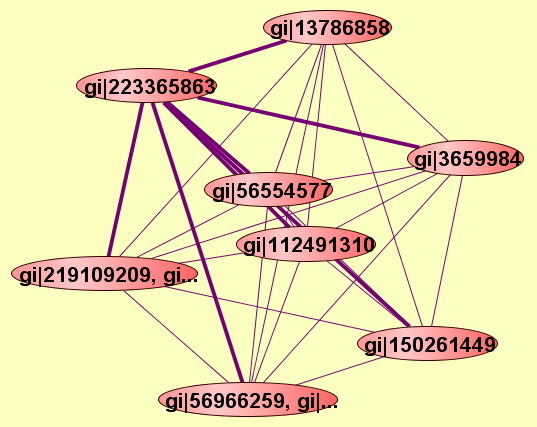
| LEGEND: |
|||
| = Same process, or one is unknown. | = Same branch, distance 4. | ||
| = Same branch, distance 1. | = Same branch, distance 5. | ||
| = Same branch, distance 2. | = Same branch, distance 6 or more. | ||
| = Same branch, distance 3. | = Not in same branch of GO. | ||
| Size | Notes | Members | |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 1BD2D and 3EDUA with confidence: 0.913174063456 | gi|223365863 gi|3659984 |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 2HE4A and 3EDUA with confidence: 0.987028789877 | gi|112491310 gi|223365863 |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 2Q3HA and 3EDUA with confidence: 0.944586886825 | gi|150261449 gi|223365863 |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 3EDUA and 1I1RB with confidence: 0.951412917638 | gi|13786858 gi|223365863 |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 3EDUA and 1XJDA with confidence: 0.950466398401 | gi|223365863 gi|56554577 |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 3EDUA and 1UNLA with confidence: 0.925203861092 | gi|223365863 gi|56966260, gi|... |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 3EDUA and 2ZHNA with confidence: 0.941343248698 | gi|219109210, gi... gi|223365863 |