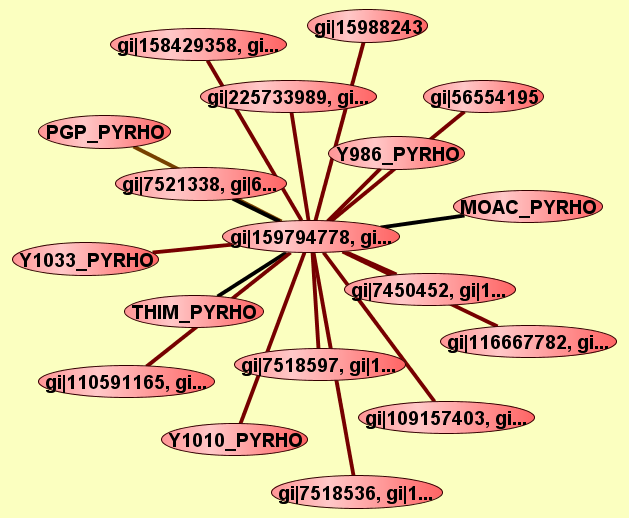
| |
Size |
Notes |
Members |
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2CVIA and 2EENA with confidence: 0.912809504602 |
gi|109157403, gi...
gi|7447797, gi|1...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 3D7AA and 2EENA with confidence: 0.923868318512 |
YA10_PYRHO, Y101...
gi|7447797, gi|1...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2ZTSA and 2EENA with confidence: 0.921684936773 |
gi|225733988, gi...
gi|7447797, gi|1...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2YX6A and 2EENA with confidence: 0.914044777858 |
gi|159795645, gi...
gi|7447797, gi|1...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2EKNA and 2EENA with confidence: 0.913114622507 |
MOAC_PYRHO
gi|7447797, gi|1...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2HQ4A and 2EENA with confidence: 0.938491945151 |
gi|116667781, gi...
gi|7447797, gi|1...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1VDMA and 2EENA with confidence: 0.924978683771 |
gi|67463846, gi|...
gi|7447797, gi|1...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2DDZA and 2EENA with confidence: 0.907468272009 |
gi|134104109, gi...
gi|7447797, gi|1...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2Z0TA and 2EENA with confidence: 0.93697666497 |
gi|7447797, gi|1...
gi|7518597, gi|1...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2P9XA and 2EENA with confidence: 0.971536800136 |
gi|158429358, gi...
gi|7447797, gi|1...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1IONA and 2EENA with confidence: 0.947720572865 |
gi|15988243
gi|7447797, gi|1...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1ULYA and 2EENA with confidence: 0.910272509386 |
gi|56554195
gi|7447797, gi|1...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1WR8A and 2EENA with confidence: 0.951077508251 |
PGP_PYRHO
gi|7447797, gi|1...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2DB0A and 2EENA with confidence: 0.92770337052 |
gi|110591164, gi...
gi|7447797, gi|1...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1V8AA and 2EENA with confidence: 0.926709410494 |
THIM_PYRHO
gi|7447797, gi|1...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2EENA and 1V6TA with confidence: 0.980748733161 |
Y986_PYRHO
gi|7447797, gi|1...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2EENA and 2HD9A with confidence: 0.969747112997 |
Y1033_PYRHO, YA3...
gi|7447797, gi|1...
|