Computational Biology
Click image to enlarge.
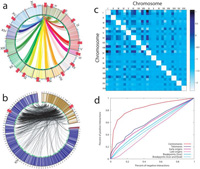
From: Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS. A three-dimensional model of the yeast genome. Nature. 2010 May 20;465(7296):363-7. Epub 2010 May 2.
On the genomics side, we are developing
several methods for characterizing large-scale genome structure.
These include methods that aggregate across a variety of functional
genomics data sets defined along the genome, as well as a 3C-like
assay that allows us to infer the 3D structure of the genome in the
nucleus.
