The Rosetta Algorithm
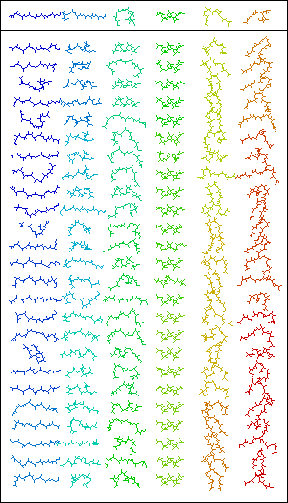
The general assumption behind Rosetta is that a short sequence of amino acids has a small number of low energy conformations. These conformations are a result of mainly local interactions and will from now on be referred to as fragments (see illustration at right).
Rosetta extracts fragments in a sliding window (i.e., 1-9, 2-10, 3-11) of 9 amino acids from the protein data bank. Rosetta predicts the unknown protein structure by assembling the fragments. After each fragment insertion, Rosetta minimizes the structure's energy.
The potential used in Rosetta tries to capture multiple features seen in experimentally determined protein structures. The potential is derived from Bayesian treatment of residue distributions in known protein structures.
Potential Terms
- environment (solvation)
- pair-wise interactions
- strand pairing
- compactness
- steric overlap
Each folding simulation results in a putative protein structure, called a decoy. We generate between 1,000 and 100,000 decoys, and we find the broadest minima by cluster analysis.
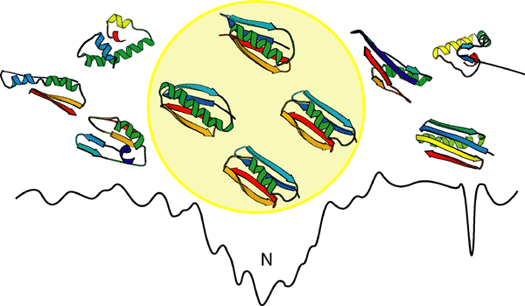
The center of the largest cluster is compared to known structures using a structure-structure comparison algorithm called Mammoth. If Mammoth can find a significant similarity between the decoy and an experimentally determined structure, it is assumed that the decoy and the matched structure belong to the same SCOP superfamily (SCOP, Structural Classification of Proteins, is a hand-curated classification of protein structures). A majority of the known SCOP superfamilies have one or two functions ascribed to them.