Sequence-Function Relationships
Click image to enlarge.
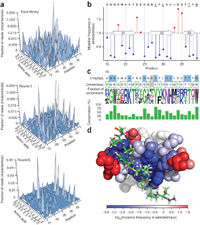
From: Fowler DM, Araya CL, Fleishman SJ, Kellogg EH, Stephany JJ, Baker D, Fields S. High-resolution mapping of protein sequence-function relationships. Nat Methods. 2010 Sep;7(9):741-6.
Deep mutational scanning can be applied to many protein activities. We would be interested in collaborations that seek
to apply this approach to biochemical activities other than protein binding and to in vivo assessments of protein properties.
We would also like to work on novel computational approaches that use these high throughput DNA data sets to model such aspects
of protein function as catalysis, interaction, stability, folding, and inhibition.