| diagram | 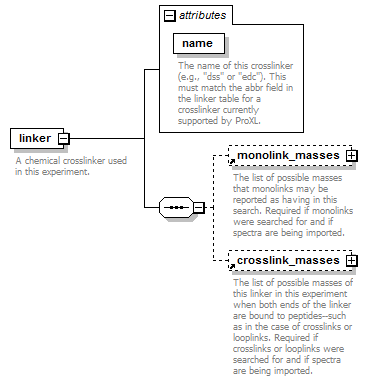 |
||||||||||||||
| properties |
|
||||||||||||||
| children | monolink_masses crosslink_masses | ||||||||||||||
| used by |
|
||||||||||||||
| attributes |
|
||||||||||||||
| annotation |
|
||||||||||||||
| source | <xs:element name="linker"> <xs:annotation> <xs:documentation>A chemical crosslinker used in this experiment.</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element ref="monolink_masses" minOccurs="0"> <xs:annotation> <xs:documentation>The list of possible masses that monolinks may be reported as having in this search. Required if monolinks were searched for and if spectra are being imported.</xs:documentation> </xs:annotation> </xs:element> <xs:element ref="crosslink_masses" minOccurs="0"> <xs:annotation> <xs:documentation>The list of possible masses of this linker in this experiment when both ends of the linker are bound to peptides--such as in the case of crosslinks or looplinks. Required if crosslinks or looplinks were searched for and if spectra are being imported.</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> <xs:attribute name="name" type="xs:string" use="required"> <xs:annotation> <xs:documentation>The name of this crosslinker (e.g., "dss" or "edc"). This must match the abbr field in the linker table for a crosslinker currently supported by ProXL.</xs:documentation> </xs:annotation> </xs:attribute> </xs:complexType> </xs:element> |
attribute linker/@name
| type | xs:string | ||
| properties |
|
||
| annotation |
|
||
| source | <xs:attribute name="name" type="xs:string" use="required"> <xs:annotation> <xs:documentation>The name of this crosslinker (e.g., "dss" or "edc"). This must match the abbr field in the linker table for a crosslinker currently supported by ProXL.</xs:documentation> </xs:annotation> </xs:attribute> |
XML Schema documentation generated by XMLSpy Schema Editor http://www.altova.com/xmlspy