| diagram | 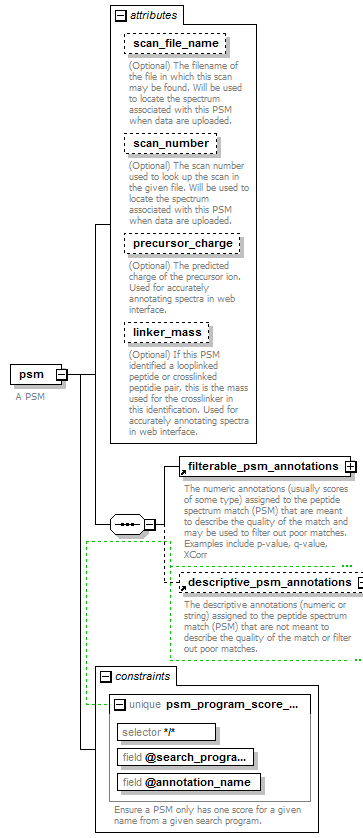 |
||||||||||||||||||||||||||||||||||||||
| properties |
|
||||||||||||||||||||||||||||||||||||||
| children | filterable_psm_annotations descriptive_psm_annotations | ||||||||||||||||||||||||||||||||||||||
| used by |
|
||||||||||||||||||||||||||||||||||||||
| attributes |
|
||||||||||||||||||||||||||||||||||||||
| identity constraints |
|
||||||||||||||||||||||||||||||||||||||
| annotation |
|
||||||||||||||||||||||||||||||||||||||
| source | <xs:element name="psm"> <xs:annotation> <xs:documentation>A PSM</xs:documentation> </xs:annotation> <xs:complexType> <xs:sequence> <xs:element ref="filterable_psm_annotations"/> <xs:element ref="descriptive_psm_annotations" minOccurs="0"> <xs:annotation> <xs:documentation>The descriptive annotations (numeric or string) assigned to the peptide spectrum match (PSM) that are not meant to describe the quality of the match or filter out poor matches.</xs:documentation> </xs:annotation> </xs:element> </xs:sequence> <xs:attribute name="scan_file_name" type="xs:string"> <xs:annotation> <xs:documentation>(Optional) The filename of the file in which this scan may be found. Will be used to locate the spectrum associated with this PSM when data are uploaded.</xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="scan_number" type="xs:positiveInteger"> <xs:annotation> <xs:documentation>(Optional) The scan number used to look up the scan in the given file. Will be used to locate the spectrum associated with this PSM when data are uploaded.</xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="precursor_charge" type="xs:positiveInteger"> <xs:annotation> <xs:documentation>(Optional) The predicted charge of the precursor ion. Used for accurately annotating spectra in web interface.</xs:documentation> </xs:annotation> </xs:attribute> <xs:attribute name="linker_mass" type="mass"> <xs:annotation> <xs:documentation>(Optional) If this PSM identified a looplinked peptide or crosslinked peptidie pair, this is the mass used for the crosslinker in this identification. Used for accurately annotating spectra in web interface.</xs:documentation> </xs:annotation> </xs:attribute> </xs:complexType> <xs:unique name="psm_program_score_unique"> <xs:annotation> <xs:documentation>Ensure a PSM only has one score for a given name from a given search program.</xs:documentation> </xs:annotation> <xs:selector xpath="*/*"/> <xs:field xpath="@search_program"/> <xs:field xpath="@annotation_name"/> </xs:unique> </xs:element> |
attribute psm/@scan_file_name
| type | xs:string | ||
| annotation |
|
||
| source | <xs:attribute name="scan_file_name" type="xs:string"> <xs:annotation> <xs:documentation>(Optional) The filename of the file in which this scan may be found. Will be used to locate the spectrum associated with this PSM when data are uploaded.</xs:documentation> </xs:annotation> </xs:attribute> |
attribute psm/@scan_number
| type | xs:positiveInteger | ||
| annotation |
|
||
| source | <xs:attribute name="scan_number" type="xs:positiveInteger"> <xs:annotation> <xs:documentation>(Optional) The scan number used to look up the scan in the given file. Will be used to locate the spectrum associated with this PSM when data are uploaded.</xs:documentation> </xs:annotation> </xs:attribute> |
attribute psm/@precursor_charge
| type | xs:positiveInteger | ||
| annotation |
|
||
| source | <xs:attribute name="precursor_charge" type="xs:positiveInteger"> <xs:annotation> <xs:documentation>(Optional) The predicted charge of the precursor ion. Used for accurately annotating spectra in web interface.</xs:documentation> </xs:annotation> </xs:attribute> |
attribute psm/@linker_mass
| type | mass | ||
| annotation |
|
||
| source | <xs:attribute name="linker_mass" type="mass"> <xs:annotation> <xs:documentation>(Optional) If this PSM identified a looplinked peptide or crosslinked peptidie pair, this is the mass used for the crosslinker in this identification. Used for accurately annotating spectra in web interface.</xs:documentation> </xs:annotation> </xs:attribute> |
XML Schema documentation generated by XMLSpy Schema Editor http://www.altova.com/xmlspy