













| General Information: |
|
| Name(s) found: |
gi|42571109
[NCBI NR]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 792 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAVSLPLFQF ILFLLLLLLS RTVYAYLIGV GSYDITGPAA DVNMMGYANS DQIASGIHFR 60
61 LRARAFIVAE PQGNRVVFVN LDACMASQIV TIKVLERLKA RYGELYTEKN VAISGIHTHA 120
121 GPGGYLQYVT YIVTSLGFVR QSFDVVVNGI EQSIVQAHES LRPGSAFVNK GDLLDAGVNR 180
181 SPSSYLNNPA AERSKYKYDV DKEMTLVKFV DSQLGPTGSF NWFATHGTSM SRTNSLISGD 240
241 NKGAAARFME DWFENGQKNS VSSRNIPRRV STIVSDFSRN RESYPFLELP NVAILYIAVA 300
301 ERLMWKVMVL FYFVSESRLL DIAATYKSSR GHSVDKSLDV KTRVRNGSKR KFVSAFCQSN 360
361 CGDVSPNTLG TFCIDTGLPC DFNHSTCNGQ NELCYGRGPG YPDEFESTRI IGEKQFKMAV 420
421 ELFNKATEKL QGKIGYQHAY LDFSNLDVTV PKAGGGSETV KTCPAAMGFG FAAGTTDGPG 480
481 AFDFKQGDDQ GNVFWRLVRN VLRTPGPEQV QCQKPKPILL DTGEMKEPYD WAPSILPIQI 540
541 LRIGQLVILS VPGEFTTMAG RRLRDAIKSF LISSDPKEFS NNMHVVIAGL TNTYSQYIAT 600
601 FEEYEVQRYE GASTLYGRHT LTAYIQEFKK LATALVNGLT LPRGPQPPDL LDKQISLLSP 660
661 VVVDSTPLGV KFGDVKADVP PKSTFRRGQQ VNATFWSGCP RNDLMTEGSF AVVETLREGG 720
721 KWAPVYDDDD FSLKFKWSRP AKLSSESQAT IEWRVPESAV AGVYRIRHYG ASKSLFGSIS 780
781 SFSGSSSAFV VV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
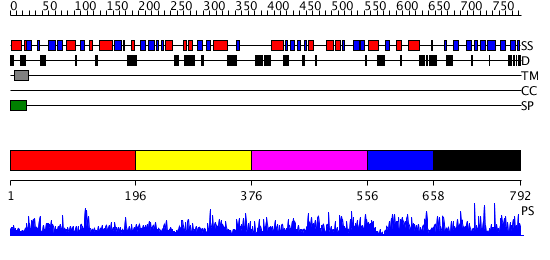
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..195] | 1.058951 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [196..375] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [376..555] | 1.042972 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [556..657] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [658..792] | 5.045954 | View MSA. Confident ab initio structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|