













| General Information: |
|
| Name(s) found: |
CAP1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 635 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytosol [IDA] plasma membrane [IDA] |
| Biological Process: |
clathrin coat assembly
[IEA]
|
| Molecular Function: |
clathrin binding
[IEA]
binding [ISS] phospholipid binding [IEA] phosphatidylinositol binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALSMRKAIG VVKDQTSIGI AKVASNMAPD LEVAIVKATS HDDDQSSDKY IREILSLTSL 60
61 SRGYVHACVT SVSRRLKKTR DWIVALKALM LVHRLLNEGD PLFQEEILYA TRRGTRILNM 120
121 SDFRDEAHSS SWDHSAFVRT YASYLDQRLE LALFERRGRN GGGSSSSHQS NGDDGYNRSR 180
181 DDFRSPPPRT YDYETGNGFG MPKRSRSFGD VNEIGAREEK KSVTPLREMT PERIFGKMGH 240
241 LQRLLDRFLS CRPTGLAKNS RMILIAMYPV VKESFRLYAD ICEVLAVLLD KFFDMEYTDC 300
301 VKAFDAYASA AKQIDELIAF YHWCKDTGVA RSSEYPEVQR ITSKLLETLE EFVRDRAKRA 360
361 KSPERKEIEA PPAPAPPVEE PVDMNEIKAL PPPENHTPPP PPAPEPKPQQ PQVTDDLVNL 420
421 REDDVSGDDQ GNKFALALFA GPPANNGKWE AFSSDNNVTS AWQNPAAELG KADWELALVE 480
481 TASNLEHQKA AMGGGLDPLL LNGMYDQGAV RQHVSTSELT GGSSSSVALP LPGKVNSHIL 540
541 ALPAPDGTVQ KVNQDPFAAS LTIPPPSYVQ MAEMDKKQYL LTQEQQLWQQ YQQEGMRGQA 600
601 SLAKMNTAQT AMPYGMPPVN GMGPSPMGYY YNNPY |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
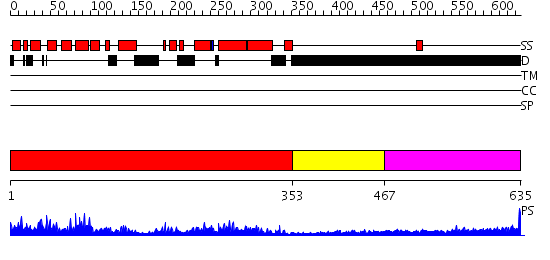
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..352] | 93.522879 | Clathrin assembly lymphoid myeloid leukaemia protein, Calm | |
| 2 | View Details | [353..466] | 1.263996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [467..635] | 8.69897 | Sec24 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)