













| General Information: |
|
| Name(s) found: |
U2A2B_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 589 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
mRNA processing
[IEA]
|
| Molecular Function: |
nucleotide binding
[IEA]
nucleic acid binding [IEA] RNA binding [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMSYEGNGDG VAISTENHNE NYISLESSPF HEDSKSRESH DLKKDSSKIS EKDNENGRDK 60
61 DGNKDRDREK DRDREKSRDR DREKSRDRDR DRERSKDRQR DRHHRDRHRD RSRERSEKRD 120
121 DLDDDHHRRS RDRDRRRSRD RDREVRHRRR SRSRSRSRSE RRSRSEHRHK SEHRSRSRSR 180
181 SRSKSKRRSG FDMAPPDMLA ATAVAAAGQV PSVPTTATIP GMFSNMFPMV PGQQLGALPV 240
241 LPVQAMTQQA TRHARRVYVG GLPPTANEQS VSTFFSQVMS AIGGNTAGPG DAVVNVYINH 300
301 EKKFAFVEMR SVEEASNAMA LDGIILEGVP VKVRRPTDYN PSLAATLGPS QPNPNLNLGA 360
361 VGLSSGSTGG LEGPDRIFVG GLPYYFTEVQ IRELLESFGP LRGFNLVKDR ETGNSKGYAF 420
421 CVYQDPSVTD IACAALNGIK MGDKTLTVRR AIQGAIQPKP EQEEVLLYAQ QQIALQRLMF 480
481 QPGGTPTKIV CLTQVVTADD LRDDEEYAEI MEDMRQEGGK FGNLVNVVIP RPNPDHDPTP 540
541 GVGKVFLEYA DVDGSSKARS GMNGRKFGGN QVVAVYYPED KYAQGDYED |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
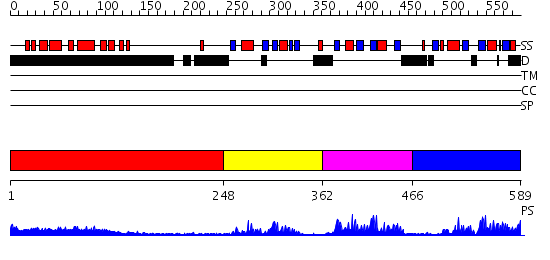
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..247] | 13.168996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [248..361] | 33.522879 | No description for 2g4bA was found. | |
| 3 | View Details | [362..465] | 20.221849 | Splicing factor U2AF 65 KDa subunit | |
| 4 | View Details | [466..589] | 15.30103 | Splicing factor U2AF 65 KDa subunit |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)