













| General Information: |
|
| Name(s) found: |
gi|162318448
[NCBI NR]
gi|162318152 [NCBI NR] |
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | synthetic construct |
| Length: | 837 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNHLEGSAEV EVTDEAAGGE VNESVEADLE HPEVEEEQQQ PPQQQHYVGR HQRGRALEDL 60
61 RAQLGQEEEE RGECLARSAS TESGFHNHTD TAEGDVIAAA RDGYDAERAQ DPEDESAYAV 120
121 QYRPEAEEYT EQAEAEHAEA THRRALPNHL HFHSLEHEEA MNAAYSGYVY THRLFHRGED 180
181 EPYSEPYADY GGLQEHVYEE IGDAPELDAR DGLRLYEQER DEAAAYRQEA LGARLHHYDE 240
241 RSDGESDSPE KEAEFAPYPR MDSYEQEEDI DQIVAEVKQS MSSQSLDKAA EDMPEAEQDL 300
301 ERPPTPAGGR PDSPGLQAPA GQQRAVGPAG GGEAGQRYSK EKRDAISLAI KDIKEAIEEV 360
361 KTRTIRSPYT PDEPKEPIWV MRQDISPTRD CDDQRPMDGD SPSPGSSSPL GAESSSTSLH 420
421 PSDPVEASTN KESRKSLASF PTYVEVPGPC DPEDLIDGII FAANYLGSTQ LLSDKTPSKN 480
481 VRMMQAQEAV SRIKMAQKLA KSRKKAPEGE SQPMTEVDLF ISTQRIKVLN ADTQETMMDH 540
541 PLRTISYIAD IGNIVVLMAR RRMPRSNSQE NVEASHPSQD GKRQYKMICH VFESEDAQLI 600
601 AQSIGQAFSV AYQEFLRANG INPEDLSQKE YSDLLNTQDM YNDDLIHFSK SENCKDVFIE 660
661 KQKGEILGVV IVESGWGSIL PTVIIANMMH GGPAEKSGKL NIGDQIMSIN GTSLVGLPLS 720
721 TCQSIIKGLK NQSRVKLNIV RCPPVTTVLI RRPDLRYQLG FSVQNGIICS LMRGGIAERG 780
781 GVRVGHRIIE INGQSVVATP HEKIVHILSN AVGEIHMKTM PAAMYRLLTA QEQPVYI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
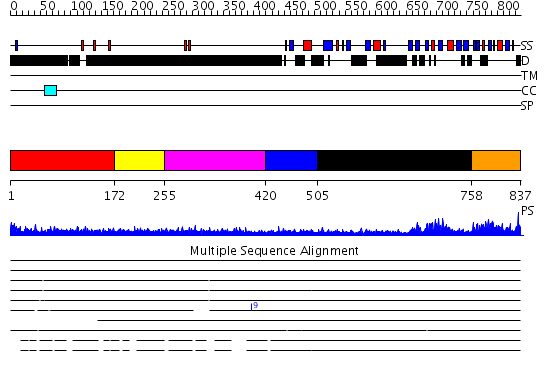
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..171] | 1.233992 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [172..254] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [255..419] | 3.251996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [420..504] | 0.96 | Coupling of Folding and Binding in the PTB Domain of the Signaling Protein Shc | |
| 5 | View Details | [505..757] | 7.221849 | Structural analysis of DegS, a stress sensor of the bacterial periplasm | |
| 6 | View Details | [758..837] | 64.39794 | Auto-inhibition Mechanism of X11s/Mints Family Scaffold Proteins Revealed by the Closed Conformation of the Tandem PDZ Domains |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)