













| General Information: |
|
| Name(s) found: |
gi|24375423
[NCBI NR]
gi|24350264 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 808 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
RNA processing
[ISS |
| Molecular Function: |
ribonuclease R activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIKDPHFERE QDKYENPIPS REYIIEYLRS QKSPITRDSI AAALQIHDEE QLEALRRRLR 60
61 AMERDGELVF TRGQSYGLPE KMDLISGTVL GHKEGYGFFK PDEGGDDLFI SNRDMLMYFH 120
121 GDKVLAQKVG MDRKGRREAR IVRLIQPRSA AIVGRFHVDS GMAFVIADDK RITQEILIAS 180
181 EDRNGARQGD VVVVELTRRP GRFVKAAGKV TEVLGKQMAP GMEIEIALRN YDLPHTWSAA 240
241 IEKKLRRIPD EVTESDKVGR VDLRSLPLVT IDGEDARDFD DAVYAEVKPS GGWRLWVAIA 300
301 DVSYYVRTDS ALDTEARARG NSVYFPSQVI PMLPEKISNG LCSLNPHVDR LCMVAEMTIS 360
361 ARGKLSGYKF YPAVMHSHAR FTYTQVAAML EGGPIAPEHE ALFVHLQCLQ SLYLALDEQR 420
421 AERGAIAFET IETQFIFNDQ RKIDKIVPRA RNQAHKIIEE CMILANVSAA KFVKKHKGEI 480
481 LYRVHESPSE QKLANFKEFL AERGLSMGGG LEPTPADYQN VMLQIADRPD AELIQVMLLR 540
541 SMRQAIYTPD NEGHFGLALE EYAHFTSPIR RYPDLVLHRV IRYLLAKEKG EANEKWTPDG 600
601 GYHYQLDELD QLGEECSTTE RRADEATRDV SDWLKCEFMQ DHVGDTFEAV IASVTNFGLF 660
661 VRLNDLFIDG LVHISSLGSD YYQFDPMRQR LIGEHTGQVY QVGDPVTVKV AAVNLDDRQI 720
721 DLMMLGDSGK GGRRKAAPSR DKPMTARERV NREGAKMAKT AKSSGAKSKA GSGKAASKSK 780
781 SKAGTKPKKS VKDTAKKPKA AKRSTRKK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
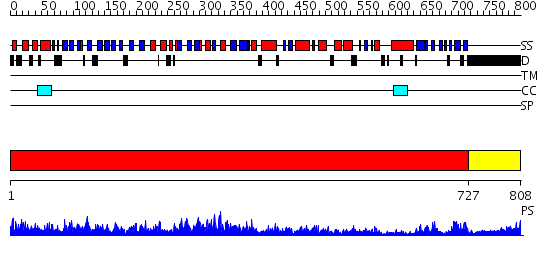
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..726] | 134.0 | No description for 2ix0A was found. | |
| 2 | View Details | [727..808] | 34.09691 | No description for 2oceA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)