













| General Information: |
|
| Name(s) found: |
gi|50924552
[NCBI NR]
gi|32482927 [NCBI NR] gi|115458582 [NCBI NR] gi|113564462 [NCBI NR] |
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 652 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQRQGRHLER SGSKRALDAG GGGGDDDDRA PKRPRVPALA SVIVEALKVD SLQKLCSSLE 60
61 PILRRVVSEE VERALAKLGP AATPARIQGR SSPKRIEGPS GINLQLQFRS RLSLPLFTGG 120
121 KVEGEQGAAI HVVLLDANTG RVVTSGPESF AKLDVLVLEG DFNKEQDEDW TEEEFENHIV 180
181 KEREGKRPLL TGDLQVTLKE GVGTIGELIF TDNSSWIRSR KFRLGLRVSS GFCEGVRVKE 240
241 AKTEAFTVKD HRGELYKKHY PPALKDDVWR LEKIGKDGAF HKKLNSNGIY TVEHFLQLLV 300
301 RDQQKLRTIL GSNMSNKMWE SLVEHAKTCV LSGKHYIYYS SDARSVGAIF NNIYEFTGLI 360
361 ADDQYISAEN LSENQRLFAD TLVKQAYDDW INVVEYDGKE LLRFKQKKKS VTTRSDTAKA 420
421 STSYPSSYGS THSHKQLTGG PVNIEQSSMS SMSEDGTRNM SNGSQAARYA ANPQDISQSI 480
481 TMPYDMSSLR PEEQFAGSSI QTQASRSSNM LALGPTQQQN FEFSALGQSM QPSPLNPFDD 540
541 WSRLQENRGG VDDYLMEEIR VRSHEILENE EDMQQMLRIL SMGGSSANMN HGDGFSPFMP 600
601 SPAPAFNYED DRARPSGKAV VGWLKIKAAM RWGIFVRKKA AERRAQLVEL ED |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
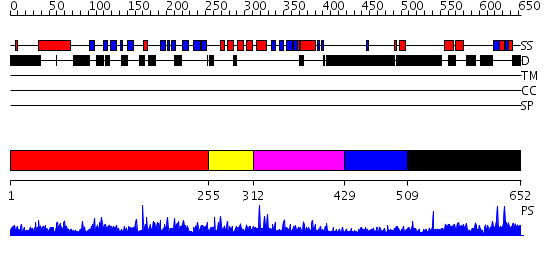
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..254] | 1.039962 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [255..311] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [312..428] | 2.044975 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [429..508] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [509..652] | 2.018956 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)