













| General Information: |
|
| Name(s) found: |
gi|16760609
[NCBI NR]
|
| Description(s) found:
Found 3 descriptions. SHOW ALL |
|
| Organism: | Salmonella enterica subsp. enterica serovar Typhi str. CT18 |
| Length: | 644 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNIFDHYRQR YEAAKDEEFT LQDFLTICRQ DRSAYANAAE RLLMAIGEPN MVDTAQEPRL 60
61 SRLFSNRVIA RYPAFEEFYG MEDAIEQIVS YLKHAAQGLE EKKQILYLLG PVGGGKSSLA 120
121 ERLKSLMQRV PIYVLSANGE RSPVNDHPLC LFNPQEDAQI LEKEYGIPRR YLGTIMSPWA 180
181 AKHLHEFGGD ITKFRVVKVW PSILEQIAIA KTEPGDENNQ DISALVGKVD IRKLEHHAQN 240
241 DPDAYGYSGA LCRANQGIME FVEMFKAPIK VLPPLLTATQ EGNYNGTEGI SALPFNGIIL 300
301 AHSNESEWVT FRNNKNNEAF LDRVYIVKVP YCLRISEEIK IYEKLLNHSE LAHAPCAPGT 360
361 LETLSRFSTL SRLKEPENSS IYSKMRVYDG ESLKDTDPKA KSYQEYRDYA GVDEGMNGLS 420
421 TRFAFKILSR VFNFDHAEVA ANPVHLFYVL EQQIEREQFP QEQAERYLEF LKGYLIPKYA 480
481 EFIGKEIQTA YLESYSEYGQ NIFDRYVTYA DFWIQDQEYR DPDTGQLFDR ESLNAELEKI 540
541 EKPAGISNPK DFRNEIVNFV LRARANNSGR NPNWTSYEKL RTVIEKKMFS NTEELLPVIS 600
601 FNAKTSTDEQ KKHDDFVDRM MEKGYTRKQV RLLCEWYLRV RKSS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
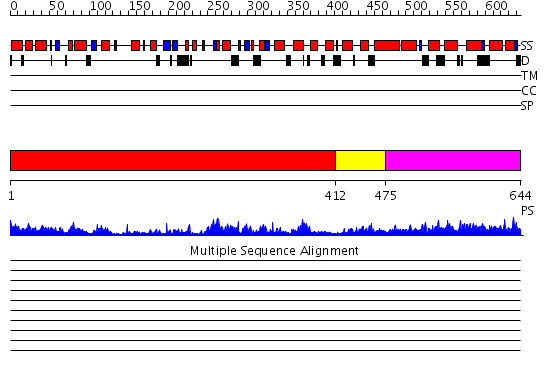
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..411] | 45.522879 | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state | |
| 2 | View Details | [412..474] | 51.69897 | Crystal structure of a sigma54-activator suggests the mechanism for the conformational switch necessary for sigma54 binding | |
| 3 | View Details | [475..644] | 72.251812 | No description for PF06798.3 was found. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)