













| General Information: |
|
| Name(s) found: |
RIPK1
[HGNC (HUGO)]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 671 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQPDMSLNVI KMKSSDFLES AELDSGGFGK VSLCFHRTQG LMIMKTVYKG PNCIEHNEAL 60
61 LEEAKMMNRL RHSRVVKLLG VIIEEGKYSL VMEYMEKGNL MHVLKAEMST PLSVKGRIIL 120
121 EIIEGMCYLH GKGVIHKDLK PENILVDNDF HIKIADLGLA SFKMWSKLNN EEHNELREVD 180
181 GTAKKNGGTL YYMAPEHLND VNAKPTEKSD VYSFAVVLWA IFANKEPYEN AICEQQLIMC 240
241 IKSGNRPDVD DITEYCPREI ISLMKLCWEA NPEARPTFPG IEEKFRPFYL SQLEESVEED 300
301 VKSLKKEYSN ENAVVKRMQS LQLDCVAVPS SRSNSATEQP GSLHSSQGLG MGPVEESWFA 360
361 PSLEHPQEEN EPSLQSKLQD EANYHLYGSR MDRQTKQQPR QNVAYNREEE RRRRVSHDPF 420
421 AQQRPYENFQ NTEGKGTAYS SAASHGNAVH QPSGLTSQPQ VLYQNNGLYS SHGFGTRPLD 480
481 PGTAGPRVWY RPIPSHMPSL HNIPVPETNY LGNTPTMPFS SLPPTDESIK YTIYNSTGIQ 540
541 IGAYNYMEIG GTSSSLLDST NTNFKEEPAA KYQAIFDNTT SLTDKHLDPI RENLGKHWKN 600
601 CARKLGFTQS QIDEIDHDYE RDGLKEKVYQ MLQKWVMREG IKGATVGKLA QALHQCSRID 660
661 LLSSLIYVSQ N |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
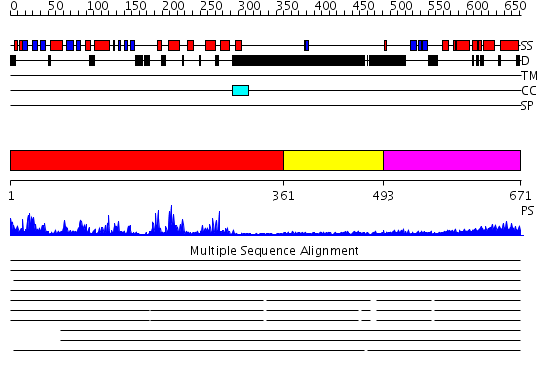
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..360] | 75.154902 | Catalytic and ubiqutin-associated domains of MARK2/PAR-1: Wild type | |
| 2 | View Details | [361..492] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [493..671] | 13.045757 | Solution structure of the death domain of nuclear matrix protein p84 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)