













| General Information: |
|
| Name(s) found: |
gi|5823166
[NCBI NR]
gi|5748520 [NCBI NR] gi|28559063 [NCBI NR] gi|119596759 [NCBI NR] gi|119596757 [NCBI NR] |
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 770 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKGDTRHLNG EEDAGGREDS ILVNGACSDQ SSDSPPILEA IRTPEIRGRR SSSRLSKREV 60
61 SSLLSYTQDL TGDGDGEDGD GSDTPVMPKL FRETRTRSES PAVRTRNNNS VSSRERHRPS 120
121 PRSTRGRQGR NHVDESPVEF PATRSLRRRA TASAGTPWPS PPSSYLTIDL TDDTEDTHGT 180
181 PQSSSTPYAR LAQDSQQGGM ESPQVEADSG DGDSSEYQDG KEFGIGDLVW GKIKGFSWWP 240
241 AMVVSWKATS KRQAMSGMRW VQWFGDGKFS EVSADKLVAL GLFSQHFNLA TFNKLVSYRK 300
301 AMYHALEKAR VRAGKTFPSS PGDSLEDQLK PMLEWAHGGF KPTGIEGLKP NNTQPENKTR 360
361 RRTADDSATS DYCPAPKRLK TNCYNNGKDR GDEDQSREQM ASDVANNKSS LEDGCLSCGR 420
421 KNPVSFHPLF EGGLCQTCRD RFLELFYMYD DDGYQSYCTV CCEGRELLLC SNTSCCRCFC 480
481 VECLEVLVGT GTAAEAKLQE PWSCYMCLPQ RCHGVLRRRK DWNVRLQAFF TSDTGLEYEA 540
541 PKLYPAIPAA RRRPIRVLSL FDGIATGYLV LKELGIKVGK YVASEVCEES IAVGTVKHEG 600
601 NIKYVNDVRN ITKKNIEEWG PFDLVIGGSP CNDLSNVNPA RKGLYEGTGR LFFEFYHLLN 660
661 YSRPKEGDDR PFFWMFENVV AMKVGDKRDI SRFLECNPVM IDAIKVSAAH RARYFWGNLP 720
721 GMNRIFGFPV HYTDVSNMGR GARQKLLGRS WSVPVIRHLF APLKDYFACE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
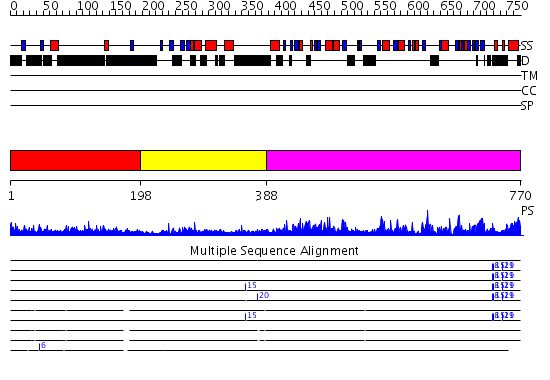
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..197] | 3.228996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [198..387] | 27.154902 | DNA methyltransferase DNMT3B | |
| 3 | View Details | [388..770] | 77.154902 | No description for 2pv0B was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)