













| General Information: |
|
| Name(s) found: |
NCASE_ORYSJ
[Swiss-Prot]
|
| Description(s) found:
Found 6 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 785 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEASSWLCYQ ARGFGSSRVW LWLLLALVLL NCSLVLSASP YLVGMGSFDI TGPAADVNMM 60
61 GYANTEQIAS GIHFRLKSRA FIVAEPNGKR VVFVNIDACM ASQIVTIKVL ERLKARYGDL 120
121 YNENNVAISG IHTHAGPGGY LQYVVYIVTS LGFVRQSFDV IVDGIEQSIV EAHNNLRPGK 180
181 IFVNKGDLLD AGVNRSPSAY LNNPAEERSK YEYNVDKEMT LIKFVDDELG PVGSFNWFAT 240
241 HGTSMSRTNS LISGDNKGAA ARFMEDWAEQ MGLPKQSAHA NSDDLRSLHK TSVLPRRVST 300
301 IIPEPNEITD DLIQLASSYE ASGGRRLAGS SITRRIRSTQ QNKPKFVSAF CQSNCGDVSP 360
361 NVLGTFCIDT NLPCDFNHST CNGKNELCYG RGPGYPDEFE STRVIGNRQF LKARDLFDSA 420
421 SEEIQGKIDY RHTYLDFSKL EVKVSTSAGG QQTVKTCPAA MGFAFAAGTT DGPGAFDFRQ 480
481 GDVKGNPFWK LVRNLLKTPG KDQVECHSPK PILLDTGEMK EPYDWAPAIL PVQMIRIGQL 540
541 VILCVPGEFT TMAGRRLRDA VKTVLTSGNS EFDKNIHVVL AGLTNSYSQY ITTFEEYQIQ 600
601 RYEGASTLYG PHTLSAYIQE FQKLAMAMIA NKEVPTNFQP PDMLDKQIGL LPGVVFDSTP 660
661 LGVKFGDVNS DVPGNSTFNK GSTVNATFYS ACPRNDLLTD GTFALVEKLD GNNNWVPVYD 720
721 DDDWSLRFKW SRPARLSSRS FATLEWTVPE DAAAGVYRLR HFGASKPMFG SVRHFTGTSR 780
781 AFAVR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
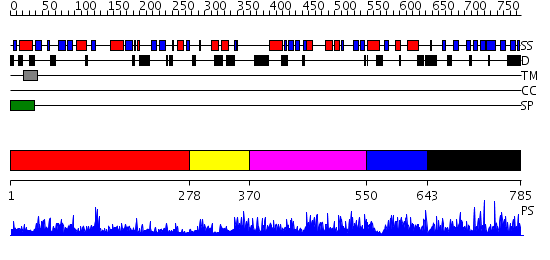
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..277] | 1.058951 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [278..369] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [370..549] | 1.039972 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [550..642] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [643..785] | 5.042954 | View MSA. No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|