













| General Information: |
|
| Name(s) found: |
CG8494-PA /
FBpp0086664
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 975 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
nervous system development
[IMP]
ubiquitin-dependent protein catabolic process [IEA] |
| Molecular Function: |
ubiquitin thiolesterase activity
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLSSNPNPH NPYVEHRNRT IHSSQGHTLA NGYAMGDPRS SRCHDKLFSG SLKSLARRED 60
61 SNASDSEGTD TAKGSGTGLV GLQNIANTCY MNSALQALSN LPPMTHYFIN CSDLVEYIAE 120
121 QSARRCKPGG LAKSYRRLMQ EIWQDVDDPK EFIAPRGILY GIRTVHPMFR GYQQHDTQEF 180
181 LRCFMDQLHE ELTEQVSMLP QTQNQPQYQS LQQQQPSETD DENDDEAAPA SLSHASESEY 240
241 DTCESSMSER SAEVLLKTEY FVTPCRTNGS NSGLPEGHSV QLQQAPLQHQ QKNASSAEQK 300
301 PIEAARSIIS DVFDGKLLSS VQCLTCDRVS TREETFQDLS LPIPNRDFLN VLHQTHSLSV 360
361 QSLNAAETSA RTNEGWLSWM WNMLRSWIYG PSVTLYDCMA SFFSADELKG DNMYSCERCN 420
421 KLRTGIKYSR VLTLPEVLCI HLKRFRNDLS YSSKISSDVY FPLEGFDMRP YIHKDCKSEV 480
481 AIYNLSSVIC HHGTVGGGHY TCFARNTLNG KWYEFDDQFV TEVSSELVQS CQAYVLFYHK 540
541 HNPQMKLVRD EAMTLSTSHP LCDSDIQFYI TREWLSRLAT FSEPGPINNQ EMLCPHGGIL 600
601 HSKADVISQI AVPISQPLWD YLYRTFGGGP AVNIIFECEI CKRAAETLSR RQQYELNEFT 660
661 KYNGLQNEFD STAIYAIAMP WLRSWQQFSR GKTHKDPGPI TNEGIAAPTE NGSATVSCVR 720
721 LGSDYAQLNA RLWRFLHNIY GGGPEIILRQ ALSDEDDAEE IEIIDQDDEC DDDEDQDLEG 780
781 EGEDEEIAAA SYQHNHSDTE SNLGIRNTPS PSPSTSPSPS LTGTRQPMET ESDLRPSSSK 840
841 SKRSRSKMKV SALRLNMRNR GKRNRSAFKQ YAEMFGAKGN YNAHSNAEPV VSEKDEKDKD 900
901 NDRTVAFPSE YSLPISIPFQ SDNFQVNGVH EKSRDKGKLR NSSKSGSTAA TKENVTLQKF 960
961 VTLREANGPS DETDI |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
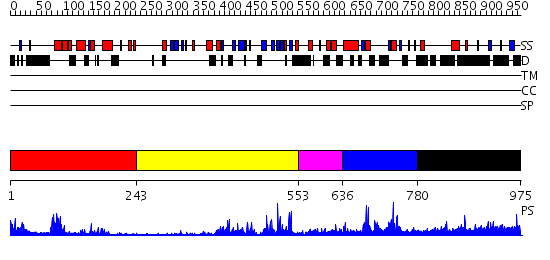
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..242] | 13.0 | Crystal structure of Ubiquitin carboxyl-terminal hydrolase 6 (yfr010w) from Saccharomyces cerevisiae at 1.74 A resolution | |
| 2 | View Details | [243..552] | 37.30103 | No description for 2gfoA was found. | |
| 3 | View Details | [553..635] | 4.88 | Solution structure of the DUSP domain of hUSP15 | |
| 4 | View Details | [636..779] | 24.221849 | Solution structure of the DUSP domain of hUSP15 | |
| 5 | View Details | [780..975] | 1.047973 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 |
|
|||||||||
| 4 |
|
|||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)