













| General Information: |
|
| Name(s) found: |
PH4alphaSG2-PA /
FBpp0085015
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 527 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum
[IEA]
procollagen-proline 4-dioxygenase complex [IC] |
| Biological Process: |
oxidation reduction
[IEA]
salivary gland morphogenesis [IMP] |
| Molecular Function: |
procollagen-proline 4-dioxygenase activity
[ISS]
L-ascorbic acid binding [IEA] oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen [IEA] iron ion binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLDRHCLYIG IFQLIIWVGV ANGEFYSSVD SMQDLAQVEE ELLNATRSYV ESQQKQLDFY 60
61 RRYVEQIKRE HEWATSQLKL DDYLGHPLHA FRLIKRLVRD WDSLIFEPIL ANNAREEFRA 120
121 FVEVLSRDLG YPDQSELQGA IKGLARLQKV YNLATSDLAD GIIGGLNYGS DLRWRECYEI 180
181 GVQLFDLGEY QRSLEWLQVA FILLRNSPRE EKDADHYLSD IREYASMANF ELGNPKKAAR 240
241 LLSQILESQP THSAQQTQKY LESRVPGKNV QETKPSWFSN YTRLCQGRRL PEERSGDPLR 300
301 CYLDGKRHAY FTLAPLQVEP VHLDPDINVY HGMLSSKQIL SIFEEADKEE MVRSAVAGSG 360
361 GEGTVRDLRV SQQTWLDYKS PVMNSVGRII QFVSGFDMAG AEHMQVANYG VGGQYEPHPD 420
421 YFEVNLPKNF EGDRISTSMF YLSDVEQGGY TVFTKLNVFL PPVKGALVMW HNLHRSLHVD 480
481 ARTLHAGCPV IVGSKRIGNI WMHSGYQEFR RPCNLTSDSY KSLAYRD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
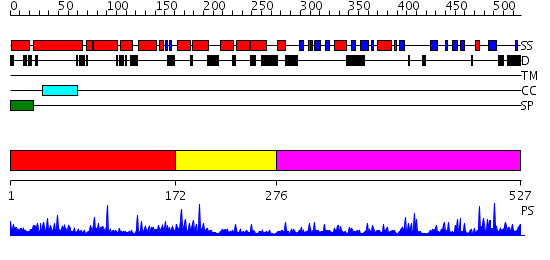
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..171] | 61.065502 | No description for PF08336.2 was found. No confident structure predictions are available. | |
| 2 | View Details | [172..275] | 17.522879 | Crystal structure of peptide-substrate-binding domain of human type I collagen prolyl 4-hydroxylase | |
| 3 | View Details | [276..527] | 22.522879 | No description for 2g19A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.41 |
Source: Reynolds et al. (2008)