













| General Information: |
|
| Name(s) found: |
sak1 /
SPAC3G9.14
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 766 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromatin [IC cytoplasm [ISS |
| Biological Process: |
positive regulation of exit from mitosis
[IMP negative regulation of transcription [IC regulation of transcription, DNA-dependent [IEA] gene-specific transcription from RNA polymerase II promoter [IC] regulation of mitotic cell cycle [IGI |
| Molecular Function: |
DNA binding
[TAS specific transcriptional repressor activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNPSDLPGQI PLSRSDMNVQ DQLDPVQRFD THFMLPQEEN FLNRPSITSE SAHPRGSDLE 60
61 QETELKRLAL EHEHYSLESL AEKLRMDHVS ANSEKFRQVF GICWLKRACE EQQDAAVQRN 120
121 QIYAHYVEIC NSLHIKPLNS ASFGKLVRLL FPSIKTRRLG MRGHSKYHYC GIKLRGQDSF 180
181 RRLRTFSDSS LSPVSCSSFP KPIPNHFEND VSSIQNTNQR VESSPASVNA AAIVRKSAVT 240
241 PSSDPYNSPP PSIPLLGSQT NLQLAPSFAA PQAHPLPSHL SQSNVPPQLS HSSVPSPAPP 300
301 RSVSQPTYFS QPMPQFSSSF VPGTSSIVPT LHPASAQEDF NLQHSLFFKL KLKFLPPHKL 360
361 PWIPSLDVSS FSLPPIDYYL NGPYDNVEAK SALMNIYSSH CITLIESVRY MHLKQFLSEI 420
421 SNFPNSLSPS LLALLSSPYF TKWIERSDTV MYREILKLLF PMTLQVVPPP VLVLLRHLAE 480
481 NLVNHISSIY ASHSSCLLQV KSETAAIFSN LLSRLLRVND TAHAAARFLA NPADRHLICN 540
541 DWERFVSTRF IVHRELMCND KEAVAALDEW YSILSTCSNP SELLDPLKDK HEASDTSMNR 600
601 VELRQIDGVL DRMADFFLEL PSRFPSCSPR MFLLCLGALQ TSVLREITVS GGEAFGALWV 660
661 IRCWVDEYMT WVAEIGGYLD DSYDELEQHH ANFHNKAGIS QSNIPPHLQE HRQSQQHFQQ 720
721 DIEALQSQQQ QQATKNSLME AAYQNAQKQK EDDYISIVFD TNGACS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
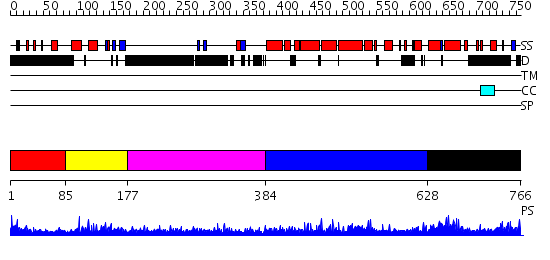
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..84] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [85..176] | 26.221849 | Class II MHC transcription factor RFX1 | |
| 3 | View Details | [177..383] | 12.226997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [384..627] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [628..766] | 2.053975 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)