













| General Information: |
|
| Name(s) found: |
SPCC584.15c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 594 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell division site
[IDA cytosol [IDA |
| Biological Process: |
regulation of signal transduction
[ISS]
|
| Molecular Function: |
calcium ion binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNKIKGGRHK SPVKLFEVRL YNAEANVIVL YGNSMDTSAR LSGIVVLSVS SPIRVKNIKL 60
61 RLSGRSFVCW ADESRHASPG NRIRRQVVQI LDKSWSFLAP NESAKVIDQG NYEYPFYYEL 120
121 PPDIPDSIEG IPGCHIIYTL TASLERATQP PTNLETALQF RVIRTIPPNS LDLMHSVSVS 180
181 DIWPLKVNYE TSIPSKVYAI GSEIPVNITL YPLLKGLDVG KVTLVLKEYC TLFITSKAYS 240
241 STCRKEFKRA LVKKTIPGLP MVDDYWQDQI MVKIPDSLGE CTQDCDLNCI RVHHKLRLSI 300
301 SLLNPDGHVS ELRNSLPLSL VISPVMFGAR PTEGVFTGDH NSYVNENILP SYDKHVFDVL 360
361 WDGIPSENPQ LQSGFTTPNL SRRNSSDFGP NSPVNIHSNP VPISGQQPSS PASNSNANFF 420
421 FGSSPQSMSS EQTDMMSPIT SPLAPFSGVT RRAARTRANS ASSVFNSQLQ PLQTDLLSPL 480
481 PSPTSSNSRL PRVRSACTLN VQELSKIPPY YEAHSAFTNV LPLDGLPRYE EATRPSSPTE 540
541 SVEIPSNTTT IAPSPVPTII APALPSTPAP PLPSHPMATR KSLSSTNLVR RGVR |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
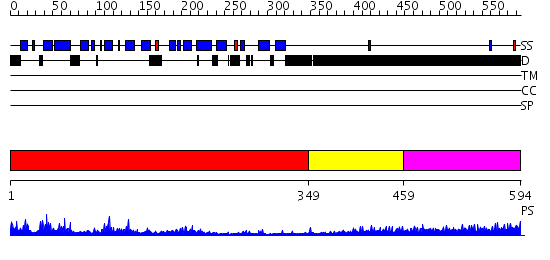
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..348] | 48.0 | Arrestin | |
| 2 | View Details | [349..458] | 0.961 | Crystal Structure of Rat Synapsin I C Domain Complexed to Ca.ATP | |
| 3 | View Details | [459..594] | 3.149996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)