













| General Information: |
|
| Name(s) found: |
gi|7522461
[NCBI NR]
gi|6013112 [NCBI NR] gi|19114191 [NCBI NR] |
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 695 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 ISSHRPEASS LLDFLVKDQK KSGDISLHTK FNLYVDDLLK KSEKGQIKKF INDIKKDLAT 60
61 ESQLPLSAPF KDESTRTMTD PQVLAYIHQS MPYQYASLYS VLTDLKIVNS DVSCKSQHIL 120
121 DCGKGPGIGA LASYSVFPTP NSVSIVEENP FLKKIIYDIH HNIYPSTSPN PTSPVTLNRL 180
181 PLGKKDSYTL VIASNKLLEM KSEKELFDYL RSLWSLVSND GGLLVLCERG TKRGFSLIQR 240
241 ARTFLLQKSK NTSDKQFNAH IVAPCPHDGR CPIDIENGVR ANICSFKQHF FLSPFSRLYV 300
301 PRSHRRSSDR SHYSYVVIQK GITRPLNNTT QRFKNDEDLL ENVNVTSPTL KNWPRIIRPP 360
361 LKRDGHVIID VCDSDARLRR NIVPKSQGKL AYRLARKSAW GDLFPLEGKV QSTSPSSKIT 420
421 KHLKDASSTY SINPPSYNKP KVERNTTADP IFVGKRFYST NRHKAFSRFA DFNSHRFPCI 480
481 FTSFSCYNCI SGTRKYSRQY SRDKFHYNQR TTIYYLVAIS IFALGLTYAA VPLYRLFCSK 540
541 TGYGGTLNTD QSRMNAERMV PRKDNKRIRV TFNGDVAGNL SWKLWPQQRE IYVLPGETAL 600
601 GFYTAENTSD HDIVGVATYN IVPGQAAVYF SKVACFCFEE QKLDAHEKVD LPVFFFIDPE 660
661 FADDPNMKDI DDILLSYTFF EARYDTNGNL LTKLN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
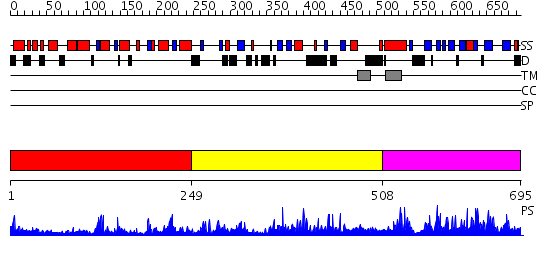
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..248] | 13.522879 | Crystal structure of aclacinomycin-10-hydroxylase (RdmB) in complex with S-adensyl-L-methionine (SAM) | |
| 2 | View Details | [249..507] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [508..695] | 68.221849 | Solution Structure of apoCox11, 30 structures |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|