













| General Information: |
|
| Name(s) found: |
Y1796_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 711 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
plastoglobule [IDA] |
| Biological Process: |
protein amino acid phosphorylation
[IEA]
|
| Molecular Function: |
ATP binding
[IEA]
protein kinase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLVVGQSLG LTLVGDGLSL RNSKINVGKS KFFSVNRRRL ARAALVQARP KEDGAAASPS 60
61 PSSRPASVVQ YRRADLADDL QAEARALGRA IDASIYSPEL IARKHGSQPF KALRRSLEIL 120
121 GALGGFALKL GIDQKQGNLE KNMKKRAIEL RRIFTRLGPT FVKLGQGLST RPDLCPPDYL 180
181 EELAELQDAL PTFPDAEAFA CIERELDLSL ETIFSSVSPE PIAAASLGQV YKAQLRYSGQ 240
241 VVAVKVQRPG IEEAIGLDFY LIRGVGKLIN KYVDFITTDV LTLIDEFACR VYQELNYVQE 300
301 AQNARRFKKL YADKADVLVP DIFWDYTSRK VLTMEWVEGT KLNEQLAIES QGLKVLDLVN 360
361 TGIQCSLRQL LEYGFFHADP HPGNLLATPD GKLAFLDFGM MSETPEEARF AIIGHVVHLV 420
421 NRDYEAMARD YYALKFLSPD VDVTPIIPAL RDFFDDALNY TVSELNFKTL VDGLGAVFYQ 480
481 YPFNVPPYYA LILRSLTVLE GLALYADPNF KVLAASYPYF AKRLLTDPNP YLRDALIELL 540
541 FKDGKFRWNR LENLLQQGSK DRDFSAKDAL QPVLKLLLDP NGEELRLLVI KEAVRVSEAI 600
601 ALGTVVDTYN SLPEFLRSLV FNGNGNGPLT MSTAELQSTL ELRDQVSRIW GLLQSSESFD 660
661 PAILQPILQV LQQPEARRLG GRVAGGVGQR LAARFLQQLL RATTPSSAPS P |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
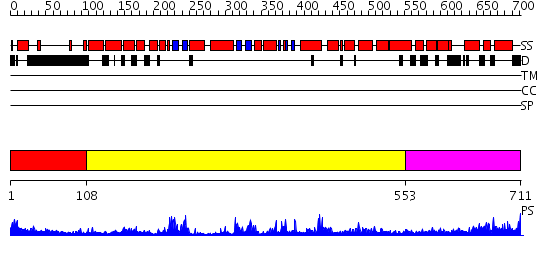
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..107] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [108..552] | 42.69897 | No description for 2acxA was found. | |
| 3 | View Details | [553..711] | 25.30103 | Ca2+-regulatory region (CLD) from soybean calcium-dependent protein kinase-alpha (CDPK) in the presence of Ca2+ and the junction domain (JD) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 | No functions predicted. | ||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.69 |
Source: Reynolds et al. (2008)