













| General Information: |
|
| Name(s) found: |
B561A_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 907 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endomembrane system
[IEA]
|
| Biological Process: |
histidine catabolic process
[IEA]
metabolic process [ISS] |
| Molecular Function: |
monooxygenase activity
[ISS]
dopamine beta-monooxygenase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCDQRPNLLG SLVLLGFFIF FVNGEECSNS SSLIGHESEF KMLQHQLRGV FTVVDDCSFR 60
61 VSRFDMLSGS EVHWWGAMSS DFDNMTNDGF VISDQKLNQT FKNSSFIVRL LGNVTWDKLG 120
121 VVSVWDLPTA SDFGHVLLSN ATESDTSKAE SPPSESNDVA PGKSNNSEPF KAPTMFDNCK 180
181 KLSDKYRLRW SLNAEKGYVD IGLEATTGLL NYMAFGWAKP NSTSNLMLNA DVVVTGIRED 240
241 GFPFADDFYI TESSVCSVKE GTATGVCPDT VYEEADSVGS SVNNTKLVYG HRIDGVSFVR 300
301 YRRPLNDSDN KFDFPVNSTE SLTVIWALGV IKPPDVINPY YLPVNHGGVE SENFGHFSLN 360
361 LSDHVDECLG PLDADNKYDQ DVIIADAHAP LVVTAGPSVH YPNPPNPSKV LYINKKEAPV 420
421 LKVERGVPVK FSIEAGHDVS FYITSDFLGG NASLRNRTET IYAGGQETHG VLSSPSELVW 480
481 APNRNTPDQL YYHSIFQEKM GWKVQVVDGG LSDMYNNSVN LDDQQVKFFW TIVGDSISIA 540
541 ARGEKKSGYL AIGFGSEMTN SYAYIGWFDR NGTGHVNTYW IDGESASAVH PTTENMTYVR 600
601 CKSEEGIITL EFTRPLKPSC SHRDRPECKN MIDPTTPLKV IWAMGAKWTD GQLTERNMHS 660
661 VTSQRPVRVM LTRGSAEADQ DLRPVLGVHG FMMFLAWGIL LPGGILSARY LKHIKGDGWF 720
721 KIHMYLQCSG LAIVFLGLLF AVAELNGFSF SSTHVKFGFT AIVLACAQPV NAWLRPAKPA 780
781 QGELISSKRL IWEYSHSIVG QSAVVVGVVA LFTGMKHLGE RNGTENVDGL NLALGLWVFL 840
841 CVVTVAYLEY RERGRRRARN LSRGNWVLGN VEEDDSIDLI DSRGGFRDKD DEDRNGGRME 900
901 IQLEPLK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
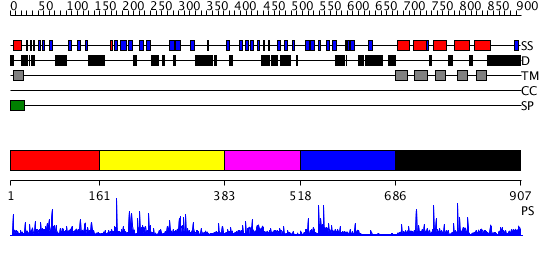
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..160] | 17.070581 | No description for PF10517.1 was found. No confident structure predictions are available. | |
| 2 | View Details | [161..382] | 11.079991 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [383..517] | 3.085996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [518..685] | 12.221849 | Cytochrome domain of cellobiose dehydrogenase | |
| 5 | View Details | [686..907] | 14.239996 | View MSA. No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|