













| General Information: |
|
| Name(s) found: |
NLP9_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 894 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: |
transcription factor activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MENPSASRDN KGFCFPDIPV EEMDGWVKNL ISEEDMFSSS STSELMNFES FASWCNSPSA 60
61 ADILFTQYGL STSQSIIPFG GLEGSYACEK RPLDCTSVPR SLSHSLDEKM LKALSLFMEF 120
121 SGEGILAQFW TPIKTGDQYM LSTCDQAYLL DSRLSGYREA SRRFTFSAEA NQCSYPGLPG 180
181 RVFISGVPEW TSNVMYYKTA EYLRMKHALD NEVRGSIAIP VLEASGSSCC AVLELVTCRE 240
241 KPNFDVEMNS VCRALQAVNL QTSTIPRRQY LSSNQKEALA EIRDVLRAVC YAHRLPLALA 300
301 WIPCSYSKGA NDELVKVYGK NSKECSLLCI EETSCYVNDM EMEGFVNACL EHYLREGQGI 360
361 VGKALISNKP SFSSDVKTFD ICEYPLVQHA RKFGLNAAVA TKLRSTFTGD NDYILEFFLP 420
421 VSMKGSSEQQ LLLDSLSGTM QRLCRTLKTV SDAESIDGTE FGSRSVEMTN LPQATVSVGS 480
481 FHTTFLDTDV NSTRSTFSNI SSNKRNEMAG SQGTLQQEIS GARRLEKKKS STEKNVSLNV 540
541 LQQYFSGSLK DAAKSLGVCP TTLKRICRQH GIMRWPSRKI NKVNRSLRKI QTVLDSVQGV 600
601 EGGLKFDSVT GEFVAVGPFI QEFGTQKSLS SHDEDALARS QGDMDEDVSV EPLEVKSHDG 660
661 GGVKLEEDVE TNHQAGPGSL KKPWTWISKQ SGLIYSDDTD IGKRSEEVNK DKEDLCVRRC 720
721 LSSVALAGDG MNTRIERGNG TVEPNHSISS SMSDSSNSSG AVLLGSSSAS LEQNWNQIRT 780
781 HNNSGESGSS STLTVKATYR EDTVRFKLDP YVVGCSQLYR EVAKRFKLQE GAFQLKYLDD 840
841 EEEWVMLVTD SDLHECFEIL NGMRKHTVKF LVRDIPNTAM GSSAGSNGYL GTGT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
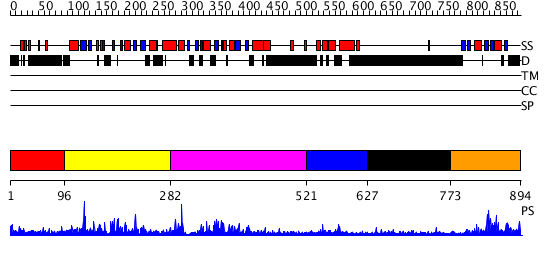
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..95] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [96..281] | 4.050971 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [282..520] | 1.081982 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [521..626] | 33.161151 | No description for PF02042.7 was found. No confident structure predictions are available. | |
| 5 | View Details | [627..772] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [773..894] | 18.09691 | Solution Structure of the PB1 domain of PKCiota |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)