













| General Information: |
|
| Name(s) found: |
gi|6850833
[NCBI NR]
gi|15233107 [NCBI NR] gi|11357716 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 566 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLLFVPNDPK SDLKLKVQTH NIKSQKYTKE GSNKPPWLNL TSKQGRSHIF EFENYPDMHA 60
61 CRDFITKALA KCEEEPNKLV VLTPAEQLSM AEFELRFKLL RENSELQKLH KQFVESKVLT 120
121 EDEFWSTRKK LLGKDSIRKS KQQMGLKSMM VSGIKPSTDG RTNRVTFNLT SEIIFQIFAE 180
181 KPAVRQAFIN YVPKKMTEKD FWTKYFRAEY LYSTKNTAVA AAEAAEDEEL AVFLKPDEIL 240
241 AQEARQKMRR VDPTLDMDAD EGDDYTHLMD HGIQRDGTND IIEPQNDQLK RSLLQDLNRH 300
301 AAVVLEGRCI IDVQSEDTRI VAEALTRAKQ VSKADGEITK DANQERLERM SRATEMEDLQ 360
361 APQNFPLAPL SIKDPRDYFE SQQGNILSEP RGAKASKRNV HEAYGLLKES ILVIRMTGLS 420
421 DPLIKPEVSF EVFSSLTRTI STAKNILGKN PQESFLDRLP KSTKDEVIHV GKLKDAMSNT 480
481 YSLLDVSYGI LLQIFFSEVV VETYSIWSSV YGFRNIIPNV NQAMKQSVQS DLRHQVSLLV 540
541 RPMQQALDAA FQHYESDLQR RTAKIT |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
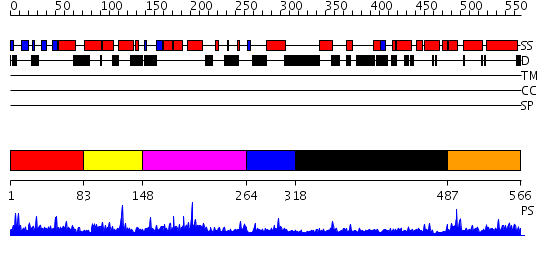
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] | 15.69897 | Solution structure of the N-terminal PH/PTB domain of the TFIIH P62 subunit | |
| 2 | View Details | [83..147] | 13.045757 | No description for 2diiA was found. | |
| 3 | View Details | [148..263] | 1.97 | Solution structure of the BSD domain of human Synapse associated protein 1 | |
| 4 | View Details | [264..317] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [318..486] | 1.034995 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [487..566] | 2.034993 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. | |||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)