













| General Information: |
|
| Name(s) found: |
GCH1_SYNY3
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Synechocystis sp. PCC 6803 |
| Length: | 234 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTIASSHSIN NGMNTGKLVN QLSSLPDRNT HNGQEVKYHP ENVELNKEKM MDAVRVMLES 60
61 VGEDPEREGL LKTPKRVAEA MQFLTQGYDQ SLEKLVNGAI FDEGHNEMVL VRDIDFFSLC 120
121 EHHMLPFMGK AHLAYIPNQK VVGLSKLARI VEMFSRRLQV QERLTRQIAE AVQEILDPQG 180
181 VAVVMEATHM CMVMRGVQKP GSWTVTSAMI GSFQNEQKTR EEFLNLIRHQ PNFY |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
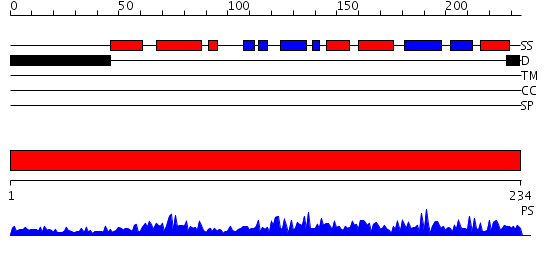
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..234] | 87.045757 | GTP cyclohydrolase I |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)